In this episode we continue our discussion of the microbiome that we started in Episode 9 and continued with Episode 37. Today we try to help you navigate the confusing field of microbiome testing companies and discuss the pros and cons of different technologies.
Examples and lessons learned from our own testing will give you an idea of how a microbiome test can help you make decisions about your health. Finally, we discuss what we think the future of microbiome testing holds.
– Richard Sprague
Long-time software executive Richard Sprague discovered his love for science through microbiome self-experimentation, studying questions like “Can I improve sleep by feeding certain gut microbes?” or “What is the impact of a gut cleanse on my gut bacteria?”
Formerly “Citizen Science in Residence” at uBiome, a biotech company, microbiomics is of particular interest to Richard because it is easy to get access to a lot of raw data that let non-specialists like him make interesting discoveries at the cutting edge of medicine and science. Richard shares his experiments and insights on his Medium Publication called Personal Science and the Microbiome and his blog.
The episode highlights, biomarkers, and links to the apps, devices and labs and everything else mentioned are below. Enjoy the show and let me know what you think in the comments!
What You’ll Learn
- Why is the microbiome interesting (5:40).
- Microbiome testing is now more accessible to the public (7:45).
- Different technologies for trying to understand your gut and what’s going on there and the pros and cons of these technologies. Technologies discussed include: Cell culture, PCR, 16S sequencing, metagenomic sequencing (9:02).
- What is the different between a different bacterial strain and a different species and why this distinction is important when analyzing your microbiome (17:40).
- Cutting edge new technologies to understand your microbiome better: transcriptomics, which looks at what genes are active, proteinomics which looks at the actual proteins and metabolomics, which analyzes metabolites (20:10).
- The reasons why the results from different labs are different (27:30).
- The different labs doing microbiome testing and compare notes on the ones they used (33:13).
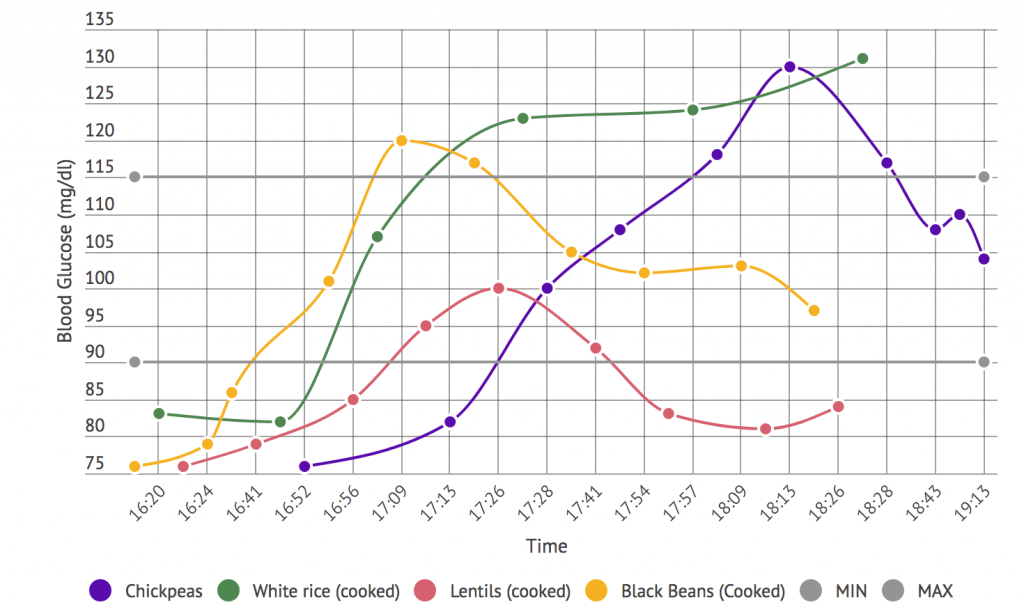
- How glucose response and the microbiome are interdependent and knowing more about your microbiome might allow you to predict your body’s glucose response to different foods (51:26).
- The labs at the bleeding edge of transcriptonomics (57:29).
- N=2 experiences with the labs used and how they interpret and compare the data they received (59:24).
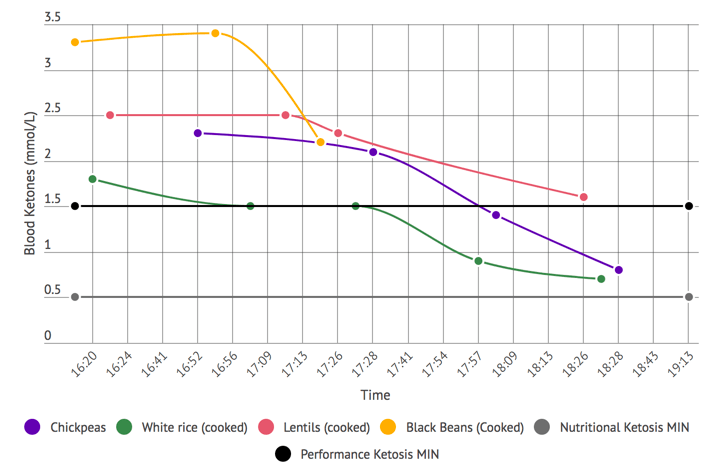
- The effects of his ketogenic diet on his microbiome (1:02:44).
- Discussion of gut microbiome diversity, day-to-day variability and individual difference in the microbiome (1:15:54).
- A self-experiment he has done to try and change is microbiome taking a probiotic and the effects of traveling and eating different foods on the microbiome (1:20:15).
- A way to change the nose microbiome using kimchi (1:22:01).
- Advantages of a varied diet over taking probiotic pills to change the microbiome (1:24:06.)
- High-level thoughts and recommendations about using different microbiome tests (1:28:34).
- Why everybody doing lab tests should try to get the raw data from the lab (1:36:30).
- Discussion of what future technologies and applications will useful to get even more information out of the body’s microbiomes (1:38:23).
- Improvements that would provide better data and insights from microbiome testing (1:41:44).
- How travel impacts the microbiome (1:47:03).
- Where to learn more about the microbiome (1:55:42).
- Information about what Richard is tracking and his interest in traditional foods and medicine (1:57:37).
Richard Sprague
- Richard’s personal website: Short life & career summaries of Richard plus articles about his microbiome self-experiments.
- Airdoc: Richard’s latest venture, a AI-based chronic disease recognition system.
- Richard on Medium.com: Richard is a top writer on health on Medium where he chronicles his near-daily microbiome experiments.
- Richard’s Twitter
Recommended Self-Experiment
Use Kefir to Change Your Microbiome
- Tool/ Tactic: Richard found a real noticeable difference in the microbiome after drinking kefir, in particular a couple of microbes that he did not have before he started drinking kefir and that he has now. Interestingly, one is associated with recovery from Crohn’s Disease. See Richard’s academic pre-print paper.
- Tracking: to track the effects of adding fermented food like kefir to your diet you need to get your gut microbiome tested before the start of the diet and several weeks or months later.
Kimchi for Sinusitis Treatment
In sinusitis sufferers the sinus microbiome is out-of-whack and the probiotic Lactobacillus Sakei is missing. L. Sakei can work as a sinusitis treatment if put into the nostrils. Kimchi is a natural source of L. Sakei. To experiment with kimchi to treat sinusitis Damien recommends the following:
- Put a teaspoon in a container with kimchi and scoop up some of the juice.
- Dip your finger into the liquid and put your fingers up both nostrils spreading the liquid.
More information on how to apply kimchi juice to treat sinusitis can be found here. The scientific paper underlying this approach is also available.
Tools & Tactics
Diet & Nutrition
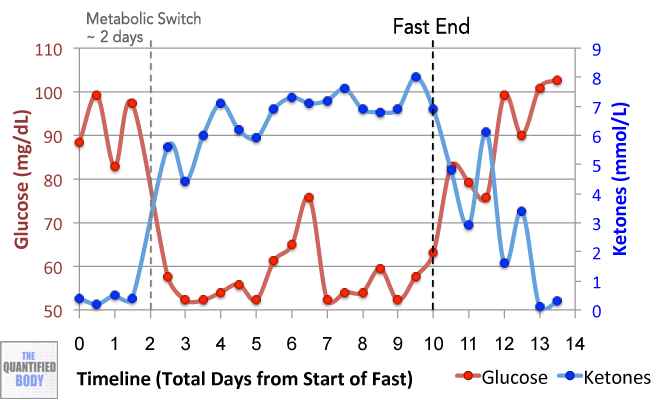
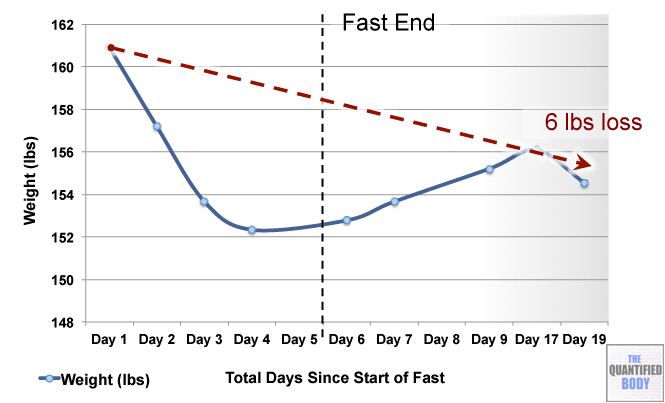
- Fasting: Fasting interventions can potentially change the microbiome. In this episode it was discussed as a tool or experiment in particular for any chronic issues/ unidentified health issues that no one knows how to solve.
Sleep
- Good sleep is essential for the body. Richard experimented with potato starch to boost his bifidobacterium levels. The result of his self-experimentation can be found in his blog. Although this approach did not work for him, other people have seen positive effects and he recommends that people with problems sleeping try potato starch.
- Damien is experimenting with three different approaches to improve his sleep:
- 10,000Lux SAD (seasonal affective disorder) light. Using this light for two hours every morning simulates strong daylight. This approach has worked for him and his theory is, that the strong light in the morning is a way of resetting his sleep cycle. SAD light use to improve sleep and prevent daytime sleepiness is discussed in this study.
- Going to bed really early also helps him to maintain a solid 7 to 7.5 hours of sleep per night. He now goes to bed by 9 pm.
- Taking a glycine supplement to reduce night wakings.1,2
Tech & Devices
- 10,000 Lux Lamp: Lamp that replicate strong sunlight. Damien has been using this in the morning to reset the circadian rhythm and as a result improve sleep quality. These lamps are designed to be used with Seasonal Affective Disorder, by providing sunlight in dark months of the year.
- Sleep Tracking Devices mentioned include:
- Zeo: A popular fitness tracker that went bankrupt due to issues with its business model.
- Fitbit: This version of the FitBit integrates sleep tracking.
- Oura Ring: OURA is a convenient wearable ring that has become popular over the last year. The company is currently participating in studies to understand the accuracy of its sleep tracking. Damien uses it to track sleep duration only – the base metric.(Note: If you’re looking at buying this discount code gives you 75 Euros off “TNBBJDQX49J”).
Tracking
Biomarkers
The biomarkers discussed in this episodes are strains or species of gut bacteria that are part of the microbiome. Tracking these biomarkers require a microbiome test.
A good best practice is to get a baseline test followed by tests over time, especially if you make changes to your diet, travel or experience health issues, to see how the microbiome tracks.
The four major groups of bacteria are Firmicutes, Actinobacteria, Proteobacteria and Bacteriodetes. Changes in the abundances of each of these groups often associate with many health conditions.
- Firmicutes and Bacteroidetes: are both key players in regulating gut metabolism, and are critical in understanding metabolism dysfunctions. See: “Diet–microbiota interactions as moderators of human metabolism” Nature 2016. The ratio of firmicutes to bacteroidetes from different lab tests was discussed, and has been discussed in the literature, but Richard is wary of relying on a single test, noting that his own ratio is highly variable day-to-day.
- Bifidobacterium also known as Lactobacillus bifidus are ubiquitous inhabitants of the gut, vagina and mouth of humans. They are found in fermented foods like yoghurt and cheese. Bifidobacteria are used in treatment as so-called probiotics, defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host”. This scientific paper published in Frontiers in Microbiology summarized the current understanding of the health benefits of Bifidobacterium.
- Spirochaete is a phylum of bacteria that contains many pathogenic species, including Borrelia species that cause Lyme disease. Testing for these pathogenic bacteria can reveal important information about one’s health. Damien put together a paper describing how one could use uBiome’s 16S rRNA microbiome sequencing as a pre-screen tool for Borrelia.
Lab Tests
Microbiome Labs Overview
With a number of different labs out there offering microbiome tests it can be difficult to decide which company to use or what the upsides and downsides may be. The table below provides an overview comparison of the different characteristics of each of the labs including.
| uBiome | AmericanGut | AtlasBiomed | DayTwo | Aperiomics | Viome | ||
|---|---|---|---|---|---|---|---|
| OFFER | Cost | $89 per test | $99 per test | £274 ($379) per test | $329 per test | $781 per test | $399/ year |
| Breadth of Testing | Gut, Mouth, Nose, Skin, Genitals | Gut | Gut + DNA (+ Metabolomics/ Blood Markers) | Gut | Gut, Blood, Urine and Oral Swabs | Gut + Metabolism (blood glucose regulation) + Body dimensions | |
| Service | N/A | N/A | N/A | Nutritionist consultation included | N/A | N/A | |
| Geographies Served | International | International | UK & Russia | US Only | International | US, UK & Canada | |
| Year Started | 2012 | 2012 | 2017 | 2017 | 2017 | 2017 | |
| TECHNOLOGY PLATFORM | Sequencing Type | 16S | 16S | 16S | Shotgun | Shotgun | RNA |
| Information Depth | From Phylum to Genus | From Phylum to Genus | From Phylum to Genus | From Phylum to Species and Strain | From Phylum to Species and Strain | From Phylum to Species and Strain | |
| Type of Information | Metagenomics (What genes are there) | Metagenomics (What genes are there) | Metagenomics (What genes are there) | Metagenomics (What genes are there) | Metagenomics (What genes are there) | Transcriptomics (What genes are expressed/ active) | |
| DATABASE SCOPE | Coverage | Bacteria | Bacteria | Bacteria | Bacteria | Bacteria, Viruses, Parasites and Fungi. | Bacteria, Virus, Parasite and Fungi |
| BENCHMARKING DATA QUALITY | Benchmark Data Quality/ Scope | Likely largest database currently | N/A | N/A | The Weizmann Institute studies included over 1000 Israeli participants on glucose regulation and the biome. Currently have study underway in U.S. | Large whole genome database covering over 37,000 microorganisms, 7500+ of which are known pathogens. | Very early stage - likely most limited currently |
| OUTPUT YOU RECEIVE | Actionable vs. Informational? | Informational | Informational: Detailed reporting. | Informational: Limited information (only family/ genus level reported) Actionable: Many specific recommendations | Recommends Actions: Rates each food according to your glycemic response | Highest level of granularity of species reported. | Recommends Actions: Rates each food |
| Transparency of Recommendations | N/A | N/A | HIGH: Includes reasing and study references for most recommendations | MEDIUM: Doesn't Explain Recommendations, but can assume comes from Weisman work | HIGH: Discussion with researchers. | LOW: No information given on which various inputs explain outputs or why | |
| Raw Data | Yes | Yes | No (planning to add?) | No (but planning to add) | Yes | No (No plans to add) |
Note: This is a high level analysis of the current technologies and labs on the market which are primarily focused on metagenomics. There are others that have yet to emerge commercially but will eventually create a broader and more complete landscape and understanding of the biome. These include metatranscriptomics, proteomics, metabolomics, and other meta data.3
Microbiome Lab Tests
- uBiome Explorer test: Richard used to work for uBiome as a citizen scientist. They use machine learning, artificial intelligence, statistical techniques, and a patented precision sequencing process based on 16S rDNA sequencing to analyze the microbes in a sample.
- American Gut: this project is run out of Rob Knight’s lab at UCSD and is one of the largest microbiome research labs in the world and the world’s largest crowd-funded citizen science project in existence. Anybody can join the project by making a donation.
- Atlas BioMed: a UK based company does DNA and microbiome testing based on 16s rDNA sequencing.
- Doctor’s Data Microbiome Testing: a clinical lab performing specialized testing.
- BioHealth GI Screens: a company providing functional laboratory testing, including testing of the gut microflora.
- Aperiomics: identifies every known bacteria, virus, parasite, and fungus in samples. Specializing in identifying pathogens and solving complex clinical infections.
- Diagnostic Solutions GI Map: microbiome testing based on PCR technology.
- Gencove: offers DNA testing to explore ancestry and tests the microbiome of the mouth.
- Arivale: tests the genome, blood, saliva, gut microbiome and is taking lifestyle into consideration.
- Viome: Analyzes the gut microbiome to help improve health, weight loss and wellbeing. Viome offers an annual plan that includes a microbiome test.
- DayTwo Microbiome Analysis: provides personalized nutrition based on the to maintain normal blood sugar levels. The company studies individual metrics and gut microbiome and translates their findings into actionable insights. Richard’s review of DayTwo can be found on Medium.
- Thryve Gut Health Test: assess gut health using 16S sequencing and provides personalized probiotics kits.
- GI Effects Comprehensive Stool Test and GI Effects Microbial Ecology Profile Test: these are tests available via Genova.
Analysis of the Different Labs
Granularity of Output from the Labs
This graph shows the level of granularity of information different labs provide to the customer in terms of number of species and genus. Some labs like Atlas Biomed only report genus level. The comparison shows that Aperiomics is able to identify more species due to the higher depth of sequencing the lab uses.
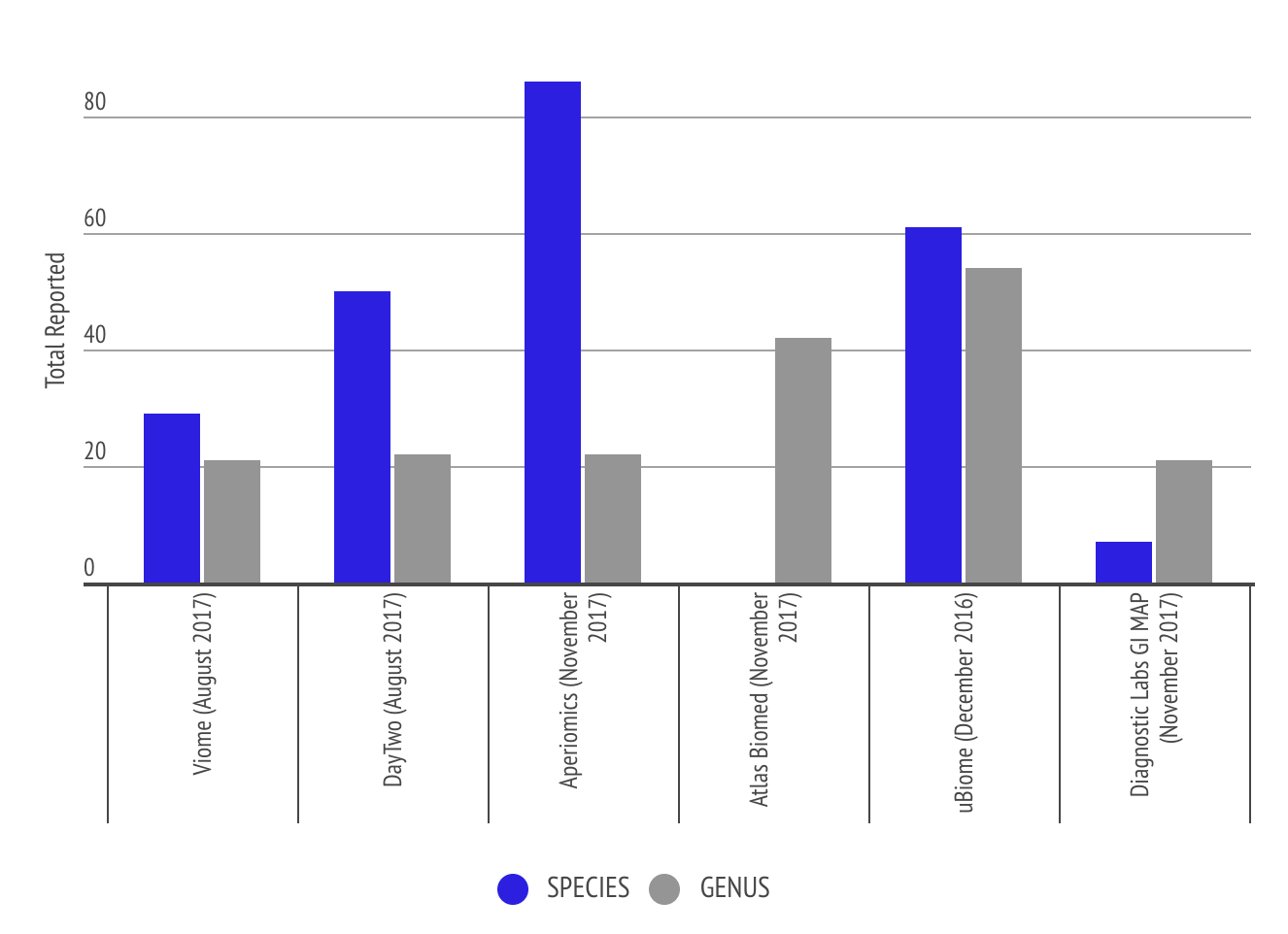
Analysis and Graphs from Richard Sprague
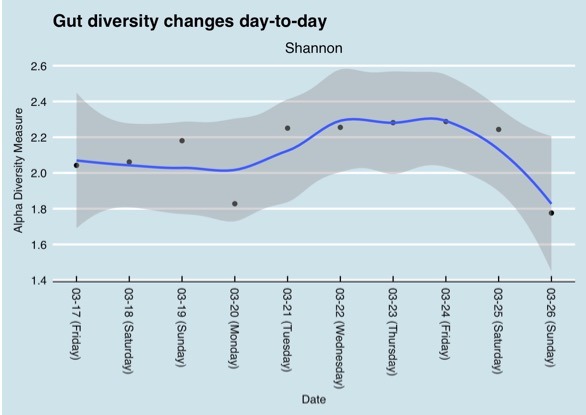
Results from different microbiome testing labs can vary by quite a bit and therefore be confusing. Some of the variety in tests results can be explained when samples are taken at different times. This graph shows gut microbiome diversity over a period of one year.
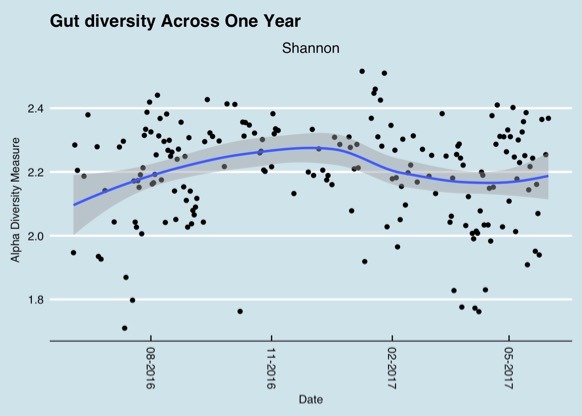
Changes in the gut microbiome over a one year period (Richard Sprague)
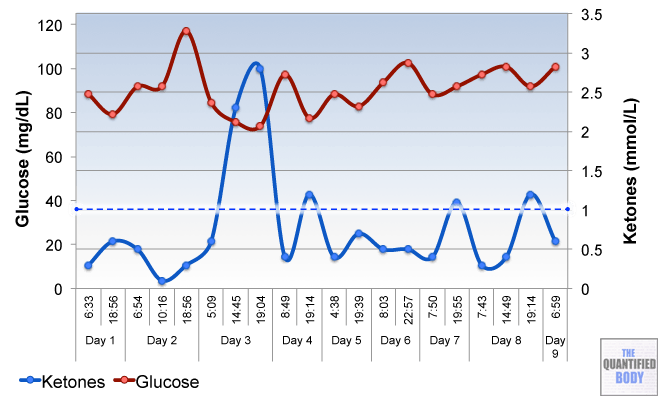
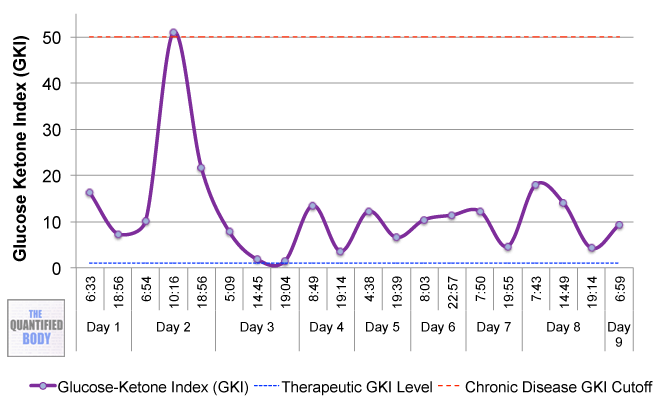
But variations can even be observed during the course of one day as the following chart shows.
But even having the same sample tested by different labs can lead to different results based on the different methods they use. To interpret data from different labs it is important to focus on the bigger picture, do the lab tests find the same type of bacteria in the same order of abundance. A chart that Richard shared emphasizes that point. The results shown in the table are from the same day, swabbed from the same tube submitted to both companies. The results are different but not extremely different. The top phyla are the same and the abundances are in the same order.
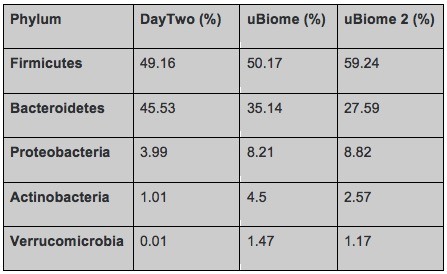
Comparison of gut bacteria phyla and relative abundance in a sample tested by Day Two and uBiome (twice) (Richard Sprague)
Other People, Books & Resources
People
- Elizabeth Bik (@MicrobiomDigest): Richard recommends following Elizabeth on Twitter. She is one of the smartest microbiome scientists he knows, and is very prolific on Twitter. She reads all the publications, and will let you know the ones that matter.
- Rob Knight (@KnightLabNews): Rob Knight is a Professor in the Department of Pediatrics at the University of California at San Diego, among many other things he is a member of the Steering Committee of the Earth Microbiome Project and a co-founder of the American Gut Project. This article in the science magazine Nature gives an overview of his work.
- Eran Segal (@segal_eran): is a computational biologist at the Weizmann Institute of Science. He has shown that there is no “One size fits all” diet, and that the very same foods can be good for some and bad for others. He is also one of the founders of the company behind the DayTwo microbiome labs. Eran was interviewed on Quantified Body with another founder of DayTwo, Lihi Segal, here.
- Chris Kresser: A functional medicine practitioner and founder of the California Center of Functional Medicine, a group of doctors that treat patients with a wide range of chronic health problems, from digestive disorders, to chronic infections, to autoimmune disease, to hypothyroidism.
Books
- The Personalized Diet: The Pioneering Program to Lose Weight and Prevent Disease: a diet book by Eran Segal and Eran Elinav that explains why one-size-fits-all diets don’t work and helps readers customize their diet to lose weight and improve health. Robert recommend it specifically because it gives suggestions for how you can test yourself using just a cheap glucose meter.
- Wired to Eat: Damien recommended this book by Robb Wolf which starts with the 30-Day Reset to help people restore normalized blood sugar levels, repair appetite regulation, and reverse insulin resistance. You can also listen to Episode 49 of this podcast for more information. This book also features standard Paleo – based recipes and meal plans for people who suffer from autoimmune diseases, as well as advice on eating a ketogenic diet.
- The Longevity Diet: Discover the New Science Behind Stem Cell Activation and Regeneration to Slow Aging, Fight Disease, and Optimize Weight: book by Valter Longo. Valter is the director of the Longevity Institute at USC in Los Angeles, and of the Program on Longevity and Cancer at IFOM (Molecular Oncology FIRC Institute) in Milan. The book describes the 5 Day Fasting Mimicking Diet which promotes longevity, overall health, and reduce excess fat.
Other
- Richard’s Favorite Books about the microbiome: a list of Richard’s favorite books and the reasons why he rates these books so highly on Medium.
- Richard’s favorite scientific papers about the microbiome: for people who want to deep dive into the science behind the microbiome.
Full Interview Transcript
(0:04:43) [Damien Blenkinsopp]: Richard, thanks so much for joining the show. It’s great to have you here.
[Richard Sprague]:My pleasure, I’m a big fan of your podcast. I’m actually a little bit humbled that you’ve asked me to come here and talk today.
[Damien Blenkinsopp]: Well you shouldn’t really be humble because you’re a real data geek when it comes to some of this stuff. So we’ve known each other for a long time because of that.
I can’t remember how we connected? Do you remember how we first connected?
[Richard Sprague]: I’m not sure either. It’s probably some quantified self thing. But I’ve been listening to your podcast since the beginning.
[Damien Blenkinsopp]: It wasn’t in person anyway, it was online. I think you must have posted you know what, I think you posted some uBiome analysis, one of the first blog posts, trying to analyze it or something and I found you on Twitter. It might be something like that.
[Richard Sprague]: It could be.
[Damien Blenkinsopp]: Okay great, so we’re going to talk about the microbiome because Richard, as I just mentioned in the intro, has been looking into this a lot. And really the first thing is just to get you guys up to speed on all of this, because it’s starting to become quite a complex question.
(0:05:40) We hear a lot about this in podcasts and health podcasts all the time. I think it’s quite a lot more complex than we generally hear. So, Richard, what do you think? What’s going on with all of this? Why is it important, and why are the labs important right now to try and quantify it?
[Richard Sprague]: You’ve had several podcast interviews with people who’ve been working in the microbiome science, but to me the way I would summarize it is that unlike genomics and genetics and your human DNA, which I find very fascinating, but there’s not a whole lot you can do to change it. Despite the fact that there are a lot of genes that are involved, there’s not a whole lot you can do if you find out that you’ve got the gene for this or that. Whereas with the microbiome you’ve got way more genes and you can change them. And I think those two things are part of the reason that I’m very excited about the microbiome.
The other thing is that partly because of that scientists are finding out all kinds of new relationships and associations between the microbiome and just about any human condition you can imagine. Everything from allergies and obesity to Alzheimer’s disease, to mental health issues like depression or schizophrenia.
There’s a relationship with the microbiome there; we don’t understand what they are, but in the last couple of years some really awesome new technology has come online that makes it possible not just to be able to go and see what the microbiome is in an individual person, but now it’s coming to the point where it’s at consumer level pricing. So that you and I can go and figure that out as well and not just wait for some scientist to go and figure it out.
[Damien Blenkinsopp]: Right. It’s actually interesting because basically since 2014 there’s been quite a few different labs coming out and these are really some of the firsts.
I mean, genetics was the first with 23andMe and players like that, but it’s one of the first areas where it’s consumer driven testing rather than coming from the medical world, and coming from physicians where they control all that stuff. But really uBiome, which was one of the first commercial players, came out and said this is going to be a consumer driven model at first.
[Richard Sprague]: Yeah.
[Damien Blenkinsopp]: Yeah. So it’s, I mean I think that’s the other reason there’s a lot of chat about it as well, because it’s more accessible to the general population.
(0:07:45) [Richard Sprague]: Yeah, that’s right. And in particular I think the 16S, I call it the Hack, made it possible to do something that people weren’t expecting to happen technologically so quickly.
Because if you think about how long and how much money it took to sequence the first human genome back in 2000. You know, that was billions of dollars and involved the cooperation of hundreds, maybe thousands, of scientists around the world.
Well, now we’re talking about at least 10 times, maybe 100 times more genes in a single human being for microbes, and they’re from thousands, maybe tens of thousands of different species. Well, how in the world would you ever sequence all of those genes? It just seems like an impossible problem.
But somebody discovered this trick several years ago that let’s you just look at 200 base pairs on one partial gene, and you can get a rough idea of what’s going on. And that just revolutionized things, because it made it possible now for people to get a hint of what all those microbes are doing.
And that just revolutionized the field. And what’s cool is like you say, since about 2014 it’s been possible for the rest of us to go and access that same kind of technology for basically under 100 dollars.
And that’s just opened up all kinds of new, interesting discoveries.
[Damien Blenkinsopp]: Yeah, yeah. So, we’ll get into why the 16S works, and how it works, in a bit.
(0:09:02) Let’s take a step back because obviously there’s quite a few different technologies out there. When you go to see physicians, when you’re using these technologies, when you’re trying to understand your gut and what’s going on, there’s a fair amount of options. And there’s different options that are being used.
So, Richard could you just give us a quick overview of what kind of technologies are being used currently?
[Richard Sprague]: The first one is culturing. And that’s been around for hundreds, arguably thousands of years, because you essentially, if you know that there’s a microbe involved and if you know which one you want, it’s well understood what kind of things they eat.
So you just take a little bit of a sample, and you put it into a Petri dish and you wait to see what happens. And, scientists know how to culture a lot of the microbes that are important, in particular the pathogens. And that’s kind of the classic way to do it; even today it’s still the gold standard. If you have some kind of medical issue where a doctor wants to confirm for certain that you have such-and-such pathogen, everybody will trust the culturing results.
So that’s kind of the first thing. The problem with culturing is that it only works on certain organisms. And they have to be alive, and it takes a while. It might take several days, or weeks in the case of some microbes.
So the next step was the development of PCR, which is if you know which microbe you’re looking for, you can put into a special machine, polymerase chain reaction, which is well understood technology that’s been around since the early 1990’s.
And they will confirm or deny whether a particular sequence of DNA base pairs are in there or not, which is another way of saying a particular microbe. And that works very quickly; that’s a few hours in some cases. And you can find out for certain whether a particular microbe is there. So the big advantage there is speed.
[Damien Blenkinsopp]: And also the accuracy, because you can really pinpoint something and if it does show up in the test, you can be sure it’s there.
Whereas even with the cultures, I think one of the issues is contamination. Because you’ve got these Petri dishes growing stuff, who knows sometimes. I’ve done some cultures in the past for different things, and I’ve been very suspect about the actual results that came out in the end. I was like, I think…
[Richard Sprague]: Yeah, you have that contamination problem with everything. The bigger issue with culturing and contamination, I think, is that sort of by definition you’re just sitting there waiting for something to happen. And sometimes it happens, sometimes it doesn’t. And, for example, if the pathogen of interest, if it somehow died on the way to the Petri dish, for no good reason, you’re not going to find it
And vice versa if the lab technician somehow exposed something or other to this or that on the way to the Petri dish then you’re going to see something you weren’t expecting.
So the next step up is, we were talking about 16S sequencing. It’s called 16S because there’s a line on the centrifuge when you take a sample and you spin it around enough, there’s a like that’s called the 16S line, which is if you skim off the goop that you find there, you will get one particular gene called the ribosomal rRNA gene. That is part of the genome that’s responsible for building the ribosome, which is an essential part of the way that all cells work.
Well, in bacteria it turns out that all bacteria use a very similar gene. We call it the 16S, the ribosomal gene. And because bacteria are all going to have that same one, in evolutionary terms it’s called conserved, throughout evolution, that it becomes possible to be able to tell the differences in bacteria based on slight variations in that gene.
The gene itself a couple thousand base pairs. But it’s one particular part of that gene called the B4 subunit that’s only, I think it’s 200 base pairs. And so if you just sequence those 200 base pairs, you got a pretty good idea of which microbe it is. Because all the different bacteria that have ever been found on Earth will have that 16S gene, and they will differ just slightly.
And if you’ve got a reference database to be able to see which one is which, and especially if you know that this came from a human gut, right there you’ve suddenly been able to eliminate having to do a gazillions of sequences. Because, sequencing something for only 200 base pairs is pretty cheap, you’re able to get the whole cost down to less than 100 dollars.
[Damien Blenkinsopp]: Yeah. So they called this hyper-variable because, I mean the interesting thing about this is that that region just varies greatly. So that’s why you’re able to identify these different genus of these sometimes species, if it happens to be a species that has more variation on that. But that’s really the key to it; it just varies so much that you’re able to identify the different things in it.
[Richard Sprague]: Yeah, and it’s pretty cool. It’s a really amazing shortcut, when you think about it.
[Damien Blenkinsopp]: Right
[Richard Sprague]: That you’re able to go from literally millions of genes, down to exactly which biome species it is. That’s pretty cool.
(0:13:44) [Damien Blenkinsopp]: And so those were the first tests that came out with the uBiome, the American Gut and some others. There’s Atlas BioMed now in Russia and the UK as well, but I’d say most of the labs are using it, the 16S. Is that the one you’ve seen because you’ve seen some others in their states, and new ones that I hadn’t come across.
[Richard Sprague]: That’s right. I mean there are lots. It’s not that hard for a lab to do 16S sequencing. In fact probably most universities do this routinely. So anybody who’s got an Illumina gene sequencer can do 16S sequencing. It’s not, the basic ideas are pretty well understood.
Also the pipeline, the software pipeline where you go from the output of the gene sequencer to actually telling you which part of the taxonomy it is. All of that stuff is available on Open Source software. Just about anyone, any feasible lab can go do it.
[Damien Blenkinsopp]: For me, when I was first getting my uBiome stuff I was trying to understand it better and I just accessed the Open Source stuff. And actually, you think it’s going to be super complicated. I didn’t do a degree in bioinformatics or anything, but actually it wasn’t that complicated.
I managed to look into, and you’ve been doing a lot of that and posting your results up online as well. That’s how you got into it. So it’s actually very accessible, which is great as well.
[Richard Sprague]: That’s right. And it’s pretty easy if you have questions to find bioinformatics experts around who will answer your questions. Because like I said, this whole technology and the basics behind it pretty well understood.
(0:15:04) So, that’s 16S. The next step up requires a lot more detail and a lot more sequencing. People call it metagenomic sequencing. And essentially what you’re doing is you’re taking the entire sample, you blow it up people say you shoot a shotgun at it and you get all these little parts flying out.
And then a computer takes, it’s almost like a big jigsaw puzzle and reassembles it. And the advantage of metagenomic sequencing is that now you’re not just looking at that one 16S rRNA gene, you’re looking at all the genes. And so it’s a lot more comprehensive.
[Damien Blenkinsopp]: And then you can get species, strain level identification.
[Richard Sprague]: That’s right.
[Damien Blenkinsopp]: Because the one thing I struggled with when I was doing a few little projects on this was sometimes if you’re unlucky and you’re trying to identify some certain species or definitely strains or even genus in some cases the 16S can’t work. It’s very difficult to get that type of level of granularity of information out of it sometimes.
[Richard Sprague]: Yeah, that’s right. And unfortunately that matters. So one of the reasons why something turns into a pathogen, it turns into a pathogen and your body isn’t able to fight it off because it may be only off one or two base pairs.
So there are versions of E-coli that are only a couple of base pairs different than the ones that are highly pathogenic. And that’s because the bacteria are able to mutate much faster than a human can. Obviously it takes us a whole lifetime before you pass on a genetic mutation.
Whereas the bacteria do this all the time. So, unfortunately most of the pathogens that you’ll see out there are just a couple of base pairs different, and you can’t tell them apart with 16S.
[Damien Blenkinsopp]: So when you say a couple of base pairs, that’s the strain level? Is that the level of strain difference?
[Richard Sprague]: That could be the strain level or the species level, it depends where on the gene the mutation happened.
(0:16:50) [Damien Blenkinsopp]: So strain for the guys at home is the absolutely tiniest, basically if you think of a human mutation, that’s kind of a strain. Do you say that’s correct Richard?
[Richard Sprague]: Yeah, the way I would describe it is that you take a dog or a wolf, both are part of the genus canine. Okay? It would matter a lot to to whether it’s a dog or a wolf at your door, it matters a lot.
So just knowing the genus didn’t help you a whole lot. The species will tell you now that it’s a dog versus a wolf. The strain would tell you that it’s a poodle or a bulldog.
[Damien Blenkinsopp]: Yeah, that’s a good example.
[Richard Sprague]: Now, there are lots of cases where it might make a big difference whether it’s a Rottweiler or…
[Damien Blenkinsopp]: A poodle, yeah.
[Richard Sprague]: Yeah. So you’ll need this kind of metagenomic sequencing to be able to tell that level of difference. And unfortunately a lot of times it matters.
(0:17:40) [Damien Blenkinsopp]: Yeah. So I had on a PCR test, just in November, fibrocholera. In other words, cholera turned up in my test. And I was looking at it like, this can’t be.
You start looking into it and you’re like, wow. I had diarrhea, stool problems, for about a week, which was very unusual, liquid diarrhea. And so I looked into this and thought, I can’t have had cholera.
And when you look into it, there’s only two specific strains of that with small modifications which cause the epidemics. The other ones, they’re dangerous, they’re not nice, they give you diarrhea for a week and it’s not nice. But it’s actually some very rare strains that come out, those are the only ones that cause the really lethal epidemics that we’ve seen in the past.
[Richard Sprague]: Could be. And in fact, and this is where it gets really complicated, it could be that the particular strain that you have will out-compete the bad guy. So having it will actually help prevent you from getting cholera.
That’s the sort of thing that happens. That’s why it’s really hard to look at the presence or absence of a particular microbe and say in isolation whether this is good or bad.
Usually it will turn out that something that’s pathogenic will have one other characteristic, which is that it is super hyper-competitive, and it will just eat up everything else and take over. And you’ll know within days, maybe hours, whether it’s bad or not.
[Damien Blenkinsopp]: Yeah.
[Richard Sprague]: So a lot of times if you just see a little bit of this or that in there, that’s just life.
[Damien Blenkinsopp]: Yeah. But I think this is really, really important because I think a lot of the people who are finding species and I think we’ve both been guilty of it too, Richard. We find a species in one of our microbiome tests, so we dig into it and we research it. Especially with the 16S lab, where it’s maybe at a higher level that it’s been identified, I think it can lead to a lot of work with no outcome there, because you’re not as sure what you’re actually dealing with.
And the best thing there is probably to escalate it, basically. If you found something in a 16S you could escalate it to a shotgun, or better PCR for the specific one that may be a concern.
[Richard Sprague]: Yeah, that’s right. And the other kind of thing to always keep in mind with all those sort of testing is that we do have a lot of data. And that’s dangerous because now suddenly you’re being flooded with a whole bunch of data, and it’s easy to overreact. Because you’ll find all kinds of things, and it takes a long time to be able to sit back and look at it a little bit more objectively and say you know what, this is just the nature of the technology. We’re at the cutting edge; we’re going to find some stuff, don’t get too excited.
(0:20:10) So, going back to the list of different ways you can measure the microbiome. One of the other areas that’s been very exciting, this is kind of where the real cutting edge is now. It’s called transcriptomics, and that’s based on the observation that just because a gene or a microbe is there, it doesn’t mean anything in and of itself.
What you really care about is whether that gene is producing the proteins that are the building blocks of life. And the way that you tell that is by the RNA that it’s producing while it’s doing all of it’s copying and transcribing these genes. So people call it transcriptomics because you’re transcribing this gene into RNA.
And there are some new tests that are coming on that let you be able to look at that. Now, that has been extremely expensive. Like I said, it’s the cutting edge and you’re talking about RNA, which is a very difficult to handle molecule; it takes special kinds of labs to be able to do that.
And what’s very exiting is that now that is becoming possible to do at consumer level pricing as well. But that’s definitly, I think most of us would agree that that’s where the future is going to be.
[Damien Blenkinsopp]: Yeah, and then after that you have proteinomics, actually looking at the proteins. Because basically what we’re talking about is the chain of events in order to create the different molecules in your body.
[Richard Sprague]: Yeah.
[Damien Blenkinsopp]: And it goes all the way down the line from genetics, transcriptomics, proteinomics, to metabolomics.
[Richard Sprague]: Metabolomics, yeah.
[Damien Blenkinsopp]: And it’s all great stuff.
[Richard Sprague]: Yeah
[Damien Blenkinsopp]: The beauty of it is one day we’ll probably have all them and actually understand what’s going on in the body.
[Richard Sprague]: Yeah, that’s right, yeah. I should also mention there are lots and lots of different tricks along the way to try to mimic what you get out of metabolomics or transcriptomics without having to do a full blood panel and that sort of stuff. One of them is called functional genomics.
For example, uBiome you can get this thing called a KEGG analysis. And that’s fairly common. That’s kind of a way to guess what sort of metabolites might be produced by this particular gene.
I don’t think it’s of super huge value. A lot of people will point to that as being evidence that such-and-such type of metabolite is present in my body. And you’ll hear that every now and then, it’s called KEGG analysis, another way to talk about it. But what I’m excited about is that now I think we’re able to move beyond that to looking more directly at what the specific thing going on in your body is.
[Damien Blenkinsopp]:With the transcriptome?
[Richard Sprague]: That’s right, yeah.
[Damien Blenkinsopp]:Yeah, I mean you can see that on uBiome, right. If anyone has a uBiome test at home they have the functional part that is displayed. Do they still have those charts, I haven’t checked for a while.
[Richard Sprague]: That’s right, yeah.
[Damien Blenkinsopp]: So that would be your KEGG analysis you’re talking about, correct?
[Richard Sprague]: That’s right, yeah.
[Damien Blenkinsopp]: And it’s things like, they’ll say you have caffeine metabolism and other things going on.
[Richard Sprague]: Or yeah, Vitamin D or this or that, yeah.
[Damien Blenkinsopp]: Yeah, yeah. I thought it was interesting because you told the story of where that came from and why we should be maybe a little conservative in thinking that that’s accurate.
[Richard Sprague]: Well it’s based on some experimental studies that were done a long time ago in Kyoto actually that’s why it’s called KEGG. It’s Kyoto something or other, EGG.
They essentially took a lot of genetic samples and they looked to see what kind of metabolites were produced. Well based on those experiments and they were carefully done experiments people are estimating when you’ve got a particular set of genes in your sample what kind of metabolites they might produce as well.
And that’s arguably better than knowing nothing at all, but I wouldn’t rely on it to be able to tell exactly how much caffeine I’m metabolizing or Vitamin D, etc.
You find a lot of this kind of stuff with genomics where somebody’s got some kind of tool, and it’s experimental. They’re just trying it out, and we’ll see how it works. And this is one of those cases. So I wouldn’t put a whole lot of stock in it.
(0:17:40) [Damien Blenkinsopp]: Yeah. Right. Great. I think another important question is why use genomics lab to understand the microbiome versus the other ones? The cultures, for example. They’re all genomics, right? The PCR, the…
[Richard Sprague]: Yeah. The biggest advantages of the genomic approaches are that it works on all of the microbes that are in the sample.
Remember with culturing, unfortunately, unless you reproduce the exact environment of your gut, which means anaerobic, no oxygen there, it’s got all the different microbes in combination and some of them are producing things that the other ones eat and need. There’s this whole community, so unless you’ve got that whole thing you can’t necessarily culture what you’re looking for.
Whereas the genomics which just says, you know what, we’re just going to look at every single gene in the whole thing. As a result, people have found that it’s well over 90 percent of all the microbes in your body can’t be cultured. We find brand new ones all the time.
[Damien Blenkinsopp]: Right. So that’s what’s going on, and that’s only been enabled by the genomics approach.
Because as you’re explaining, it’s super complicated; all the interactions between the bacteria and they rely on each other to survive.
As soon as you remove them and you’re trying to culture them or something, you remove that whole environment that they’ve been able to survive and breed in. And they need the metabolites, the things coming from the other bacteria and they’re just not there, potentially, because you kill them off.
The way that culturing works is basically you’re trying to separate out the things you’re trying to grow so that they show up in color and stuff. But by separating out and killing off the other stuff and not letting it grow, you’re basically killing off the ones that you want to grow anyway, in some cases, because they need the other bacteria.
[Richard Sprague]: Yup, that’s right. And it turns out that in a lot of interesting cases like some of the pathogens, maybe that’s good enough. But if you’re really trying to understand the whole richness of the microbiome, you’ll have to go to the genomics approaches.
[Damien Blenkinsopp]: Excellent.
[Richard Sprague]: So, now I will say, and I think we should put a big caveat in here. The genomics approach is nice to be able to get a look at all the genes that are there.
When I first started studying this, I thought, wow this is awesome, I’ll finally know what’s going on in my body. But I discovered that it’s actually much, much more complicated than it looks. As you can imagine, if you’ve got millions of organisms in a sample and you want to turn that into some useful data summary, there are a lot of steps that the lab has to go through.
And the steps are everything from the way that you happen to insert the sample into the vial, and it goes through the mail, and then how the lab tech handles it. All the way up to the bioinformatics pipeline where they’re going to process all of these numbers that come out of the sequencer and turn that into whatever taxonomy.
There are dozens of steps involved, and in any of those steps if the lab does it slightly differently than the other lab, you’re going to get different results.
[Damien Blenkinsopp]: Correct me if I’m wrong, because Richard has been at uBiome for quite a while so he’s had a closer experience with all of this. It seems like the bioinformatics pipeline, which is basically a series of calculations you’re going to make based on a database you have of references.
[Richard Sprague]: Yeah
[Damien Blenkinsopp]: And that comes from research of things saying that this piece of code means that this species, genus, exists, and so on. So you’re using a database of references in order, and you’re pushing it through this pipeline of algorithms, basically, that looks at the database checks and categorizes things. So that’s what that bioinformatics pipeline is actually doing.
And it turns out that everyone’s creating their own bioinformatics pipeline, and they’re using different databases, different reference databases
[Richard Sprague]: That’s right.
(0:27:30) [Damien Blenkinsopp]: And then we get quite different results, which is the next question I wanted to bring up. Why are we getting different results from different labs?
[Richard Sprague]: Yeah. And this is a little scary for me when I started digging into this, because I had spent a lot of time getting to know the different papers and the different labs and the different conclusions that people had come up with.
And you can put it in the show notes, but there’s a chart that I like to see that was from a publication in Science a couple of years ago where somebody actually went through and compared all the different big microbiome categorization projects and looked at just some of the common genus level microbes that they found in there. (publication referenced by Richard)
And it’s a little scary, because you look at it and you see that oh, the Human Microbiome Project says that such-and-such genus is dominant, and this one big study of like 4,000 individuals in the Netherlands found that no, that’s not the one that’s dominant, it’s a different one. And we’re talking about hundreds of thousands of individuals, so you’d think that they would all kind of average out, but that’s not the case.
And even American Gut and uBiome, if you look at their overall pictures, when they look at firmicutes versus bacteroidetes, or some of the other common ones, the results are just different. And you could say that, well maybe that’s because the type of people who send samples to uBiome are different than the ones who don’t.
But you’re talking about enough people that that’s a little bit harder to swallow. So what’s really going on is that a lab makes just one little change in, for example, how many times they PCR something before they submitted the sequencer, just one little change like that will express different levels of DNA, and then poof, you’ve got a different result.
And each of the labs if they use different reference databases, like you were saying, those references could be slightly different. If they find that a particular gene, they look it up in one reference database and it says that, oh this is bifidobacterium such-and-such. Well another lab might have called it something else.
So you just have to be a little careful. The good news is, and this is the way I look at this, if you’re going through the same lab most labs, I give them the benefit of doubt that they’re usually pretty careful. And the scientists behind this are usually pretty cautious about how they do protocols.
So you could usually trust when you submit a sample to one lab that it’s comparable to the sample the next time you submit it to the same lab. It’s just you have to be a little bit careful if you see a paper that says that they found that such-and-such microbe is associated with such-and-such condition, don’t just automatically assume that, oh my uBiome results says I have that microbe then that must mean I have such-and-such association.
[Damien Blenkinsopp]: Yeah, you could look at which lab did they use. Basically. And it’s a shame that there isn’t a standardized reference database, but it’s also the nature of the technology and the way it’s developing really.
[Richard Sprague]: That’s right, yeah.
[Damien Blenkinsopp]: Because it’s been opened up, and we have this commercial model. Which is actually enabling really the explosion of data gathering.
I don’t know how many samples, but basically there weren’t enough samples out there being collected and so on to advance science, right? So you have these commercial companies, like uBiome and so on, and they’ve made it feasible to get a large number of samples. I don’t know if you know how many samples uBiome has now, or if that’s disclosable.
[Richard Sprague]: I think the last announcement they’ve made is it’s well over a quarter million. I don’t know the exact number what they’ve announced, but it’s a lot of samples.
[Damien Blenkinsopp]: Right. And then you learn a lot from that massive data, you start the see the correlations. All the labs have, I think, questionnaires filled in as well so that they can start to see if there are some things that are related to Paleo diets, Keto diets, to antibiotics abuse. Not that many people like to abuse antibiotics in particular, but it has been done.
So I think it’s really interesting that all this data is being collected. And the nice thing, also, is that they keep the sequences, correct? This is definitely an area you’ll know more about than me, but if we wanted to run this through a different bioinformatics pipeline later, could we do that?
[Richard Sprague]: It would be tricky. Are you saying like if I submitted the same sample to uBiome and later on to someone else?
[Damien Blenkinsopp]: No I’m saying uBiome has a million samples, for example. And they have a particular bioinformatics pipeline today which says that, for example, I have a species we’ll talk about the cholera species that came up in my PCR test recently. But maybe in the five years time they’ll improve their reference database.
[Richard Sprague]:Yeah, that’s right. So, in fact, they could just go back to the shelf and look up and see your old sample and then run it through something else, and they might find something new. That’s right, yeah.
[Damien Blenkinsopp]: Right, so if they ever do decide that it’s important to change their bioinformatics pipeline, they could…just run it again.
[Richard Sprague]: Yup, you could run it again. And in fact, if you have the fast Q file, the raw output from the sequencer, it’s possible to run it through a different pipeline there as well. And if in the future somebody comes up with a better reference database, for example, it’s possible to take that same exact fast Q file and come up with a different answer.
(0:32:28) [Damien Blenkinsopp]: Well exactly. So they have all these fast Q files on a server somewhere, I’m guessing. Right? So these are the things you could run through a bioinformatics pipeline and get different answers. So that data is going to be invaluable, incredibly valuable.
[Richard Sprague]: Yeah, you’ll be able to find new insights from the old data in the future.
[Damien Blenkinsopp]: Right. Richard and I were just talking before we started this episode, some of this stuff may be challenging to get without visuals.
Whenever we’re mentioning something and it sounds complicated, we’ll probably throw a chart in there because we’ll realize that, and we’ll be like yeah, that one deserves a visual chart. So we might go over the concept relatively quickly, because we realize we’re not going to get there on audio but try and provide some visual aides in the show notes.
(0:33:13) Let’s talk about the actual labs now. What are all these labs? We’ve just kind of bounced around a few of them already, but what’s the landscape look like? It looks like it’s kind of exploding in the last few years, right? So I think uBiome and American Gut were around in 2014, and since then there’s quite a few different labs that have come out.
[Richard Sprague]: Yeah, that’s right. I’m actually curious also about you, because you’ve done more of the culturing than I have. And what kind of labs you’ve had experience with on the culturing side.
[Damien Blenkinsopp]: Yeah, so there’s basically a lot of functional medicine practitioners and hospitals in general will use the culture approach.
So I’ve done many, many different cultures over time and eventually this led me to running two different cultures; this was quite a few years after having started the Doctors Data and the BioHealth lab side-by-side, because they have different strengths and weaknesses. They’re both culture based test, and pretty consistently some things would turn up, but not necessarily on both of them.
I was working with Chris Kresser’s California Center of Functional Medicine there. And I like those guys because they’re very conservative about tests; you may have come across them as well Richard, I know they were talking to uBiome.
And they’re very conservative about their tests. They look for the studies, they look and they have a very large population of clients as well. And they’ve been running for many years. So I like the fact that they’ve been doing that for a while, and they have changed their tests over time.
And they, I think they may have moved on a little bit from these tests, but a couple of years ago when I was doing a lot of this with them they were running both of those side-by-side. That’s a little bit expensive, but it did tend to give us pretty clear…
[Richard Sprague]: So, did you submit the same exact sample to two different labs?
[Damien Blenkinsopp]: Yeah. Each time. Yeah, that was their protocol. Basically they…
[Richard Sprague]: And can I ask you, those culturing labs were they, did you have to poop in a box or did you just send a swab?
[Damien Blenkinsopp]: We used these kind of tiny vials for the uBiome, right? Where you put this really little vial, I mean basically the size of the end of your thumb. The culture labs, they’re larger; kind of three times a test tube size. They’re like a big test tube.
[Richard Sprague]: So a couple of tablespoons?
[Damien Blenkinsopp]: Yeah. And normally, actually, you have four of those for each kit. So there’s a lot of spooning and scooping that goes on for a little while into these different containers because they’ve got different assays they’re running there and they’re trying to preserve and do different things in each of those vials so they can look for different things, parasites and so on.
So it was quite a time consuming process when you’re doing that.
[Richard Sprague]: Yeah. And did you have to go to the hospital or the doctors office to do it?
[Damien Blenkinsopp]: Yeah, you do these from home. They send you the kits, and you sit on the floor scooping. I would lock myself in there for half an hour and scoop away.
[Richard Sprague]: And did the tests agree with each other? You said you submitted from the same sample.
[Damien Blenkinsopp]: Sometimes, sometimes they didn’t.
The reason they were using those in particular was because they felt they had different strengths as well. The last I heard some people feel BioHealth was a little more useful and picked up more stuff.
And again it comes back to our discussion of sensitivity, whether it’s picking up stuff. And that is the concern with a lot of physicians that it’s not picking up stuff, and it doesn’t do it reliably.
So I actually experienced this because I did many of these, over time. We were doing them every couple of months or so to see if the treatments we were doing against parasites like blastocystis hominis I had that for a while, and it’s quite a common thing but it can be a bit of an annoyance in the gut.
And we would do a protocol to get rid of it; we would retest it, it’d be gone. And we’d wait. You have to wait after your treatment, obviously, in order to let things settle down and then see if they grow back. And it would be gone for maybe two tests, and then it would come back again; it would just pop up on one of the tests.
So there’s a bit of inconsistency, and it’s a little bit worrying. For that reason you end up doing a lot of these, and they can be expensive.
[Richard Sprague]: Yeah, interesting.
I don’t know too much about Doctors Data or Biohealth.
I talked to functional medicine practitioner who used GI Effects. And that seems to be, at least in the Seattle area where I am and a lot of naturopaths, that seems to be kind of the one that most people use. The functional medicine people that I talk to are pretty positive about it, and they say that it actually produces very actionable results for treatment.
[Damien Blenkinsopp]: Yeah.
[Richard Sprague]: It seems the one to beat.
[Damien Blenkinsopp]: That was actually the first one I ever did, I think back in 2011 or something. It was MetaMetics previously and Genoma acquired that company. And MetaMetrics was very well respected as a company, so it was a good acquisition.
It came up with some stuff. And that is a combination of the culturing approach and PCR which we were talking about later, which is a genome sequencing but quite accurate. If you see something with PCR, it’s there. That’s a high probability.
[Richard Sprague]: Yeah, and I would say I’m not a doctor and please don’t trust my advice but if I did have some kind of gut issue I would want the functional doctor to use what he or she is familiar with and comfortable with, and they seem to be comfortable with this. And I would trust those results because they’ve been used for years and years and doctors have learned what work or don’t work about them.
I look at the other genomic results like the 16S and metagenomic results as being kind of cool for someone like me, and definitely worth watching for future potential. But if I were really sick, I would want to stick with what the doctors trust.
[Damien Blenkinsopp]: Yeah, exactly. And so I know some functional medicine practices have evaluated 16S based testing, and have done trials with it. But so far they’re like, this isn’t going to be good enough in terms of diagnostics, and also just the cost. Maybe it would pull out some things sometimes and be a little bit indicative of something, or just help you to explore doing a PCR with something. But they felt like the cost benefit and just the kind of time involved in getting a patient to do it wasn’t worth it at this point.
(0:39:10) [Richard Sprague]: Yeah, maybe. Now, on the other hand, there are a lot of conditions where the traditional culturing, or even the PCR approaches, can’t find out what’s wrong, and they don’t know what’s going on.
And that, I think, that’s where the place is for a little bit more experimental and you want to look at a bigger picture. And that’s where you get the 16S and metagenomic approaches because you will see a lot more.
[Damien Blenkinsopp]: Absolutely.
[Richard Sprague]: And after you’ve looked at zillions of samples the way that I have, you do start to see patterns. And you start to see when something looks anomalous, and you say, hmm. And those are the kind of things that if you’re just relying on culturing approaches you probably wouldn’t be able to see
[Damien Blenkinsopp]: Absolutely.
I’ve been really interested in the shotgun approach in particular for this, to pick up things that, as we said before, with PCR basically you have to say I want to find a poodle. You know? Or I want to find a dog in the mass of everything in the world. So you have to really know what you’re looking for, otherwise you’ll just get a negative and it costs money.
Whereas the shotgun, if you don’t know what you’re looking for but you think there’s something there it’s a good idea to do a shotgun to give you an indication. So I did a recent one, Richard and I were talking about the shotgun approach which is looking for pathogens and things like this, which is the Aperiomics, the lab test.
And I did a shotgun sample of my poop and, you know, there were a few different pathogens and some others that came up which were unknowns. A lot of them were unknowns, actually, because it’s a relatively new service; and this is where you see the bioinformatics pipeline, their reference database and so on.
They told me the benchmarks they have so far. They don’t have enough data, so there’s some interesting stuff, but there’s a lot of unknowns; we don’t know if its pathogenic or not because a lot of people have this and they’re going fine.
But I think that for me was an interesting test because it was using shotgun just to potentially pick up something interesting, and then go after it with PCR.
[Richard Sprague]: Yeah, that’s interesting. And I would love to see results that people do side-by-side if you submit the same sample to two different labs. It would be really interesting to compare that.
(0:41:12) [Damien Blenkinsopp]: Yeah, so I did that with the GI Map from Diagnostic Laboratories. Also uBiome, but unfortunately somehow that was lost, either in the post or I don’t know what happened with it. And I did Free Labs.
GI Map, we haven’t discussed, is a PCR based test. And that’s from Diagnostic Laboratories. And there’s a lot of functional medicine practitioners who are now looking at that one. Because it is PCR based, so again if you pick something up and it’s looking for quite a number of problematic bacteria, parasites, and so on, then it can be pretty useful. It’s a little bit more expensive, but that’s a good one.
So I ran that next to the Aperiomics, and I had that back. And I was trying to cross them, but nothing crossed actually.
[Richard Sprague]: Oh you didn’t find any, there was no consistency between the two?
[Damien Blenkinsopp]: No, I didn’t find the same. So I found the cholera in the GI Map. So I trust that because it’s PCR based. It didn’t turn up in the shotgun, which could be the reference database that they haven’t put that species in, or that specific strain in even. Or it could be the bioinformatics pipeline that they haven’t built out yet.
There’s so many different reasons that that might not be. But it goes back to what Richard was saying earlier, is that if you’re using different labs it’s not necessarily going to pick up the same stuff at this stage.
[Richard Sprague]: That’s interesting if they couldn’t find cholera in two different samples.
Part of it also could be if we’re talking about minute amounts, even the metagenomic approaches you’re only looking at a certain number of, you’re not looking at every single gene in there. You’re still looking at a subset of all the different genes, because you can’t sequence all gazillion of them.
The PCR approach though, you’re looking for a particular one. So you stick in some primers that will cut every single copy of DNA that has that one in there. You’d have to ask somebody who’s more knowledgeable about lab science than I am to state this more unequivocally, but when you do that you will know that the following DNA snippets came from that microbe.
Whereas with the shotgun approach, you’re going to know at a broad level, because you’ve looked at as many as you could, but you haven’t looked at every single one of them
[Damien Blenkinsopp]: Yeah.
[Richard Sprague]: And when you’re talking about minute amounts, that might make the difference.
[Damien Blenkinsopp]: Yeah, I think the nice thing about, going back to genomics, is that it will get better over time, as these databases, these bioinformatics pipelines, as each company basically gathers data and experience. And eventually, hopefully, there will be some type of collaboration. I don’t know what would be up in the future, but it would be nice if there was a way to match these together and get…
[Richard Sprague]: That would be neat, yeah. That would be neat to have a bunch of people all comparing our results from the different labs.
[Damien Blenkinsopp]: Yeah, and trying to build conversion tables or something. Something like a pool where you can convert your uBiome into your American Gut, or whatever you wanted. And it would be more comparable.
(0:44:07) [Richard Sprague]: And see how you compare, yeah. In fact, actually it’s funny because American Gut is one of the few labs that you submit the sample dry. In other words, you just put it on a Q-tip and you send it in dry. You don’t put it in a special upright vial.
[Damien Blenkinsopp]: Nothing? Okay, interesting.
[Richard Sprague]: And I asked the lab about that, because that’s kind of odd. And they know that there are certain species that when they are dry they continue to multiply. Because it’s not dead when it comes out of your body. And some of them when they’re exposed to oxygen immediately die, but some of them don’t. In fact, some of the thrive, and you get a bloom actually in some species.
And what American Gut does and they’ve written a paper about this, they’re very upfront about it they run, in their bioinformatics pipeline, they’ve already tested which species are thriving in an oxygen environment, and they filter those out. And they say oh well you collected a sample on such-and-such date, that means that this much time has passed which means that likely this much of this species has bloomed. And we’re just going to go and adjust the final result that way.
[Damien Blenkinsopp]: Whereas basically uBiome’s test and others, they’re killing all the bacteria straight away to preserve them in the state they were in the stool.
[Richard Sprague]: Yeah. And again, that’s going to be a difference in the pipeline. You’re going to get different results.
[Damien Blenkinsopp]: I mean, I can imagine. I mean that introduces basically another variable. I wonder why they didn’t decide to eliminate that.
[Richard Sprague]: Well the reason they didn’t do that is because the people at American Gut are super careful scientists, and what they care mostly about is consistency across all their different samples. They want to make sure that every single sample is conducted under the same conditions.
And they also at least in the beginning were working a lot in environments like outside the United States where maybe the collection procedure was maybe a little bit more erratic. And they just wanted it so that they could take all the different samples and treat them exactly the same way.
They’ve got a paper on this where they show, you know, that it doesn’t matter as much as you might think, but still. Yeah, it’s another area where the pipeline is going to be different.
(0:45:55) For you guys at home, just a quick reference there. I spoke to Rob Knight from the American Gut a while back, so if you wanted to know more about what he was doing there. He talks about where they got the first data and so on for that project.
(0:46:10) Okay, great. How about the 16S labs? Because you know all the 16S labs really well.
[Richard Sprague]: Yeah, well let’s talk about the 16S.
Now, first of all, I want to repeat in full transparency, I am a friend of uBiome. I’m a former employee. I’ve been a happy user of them for a long time. But I have spent time with their scientists; I trust their scientists. I think they’re pretty careful about how they put stuff in the lab.
Now that said, there are lots of other labs that I’ve worked with as well, and I’ll just go through the differences.
We talked about American Gut. I think that American Gut is scientifically they’re the most sound lab. You’ve had Rob Knight on this show, you know he’s a very smart guy, well published, extremely careful scientist, and knows everybody.
They have published a lot of results based on their American Gut cohorts, and they’ll continue publishing. They take their science very seriously. The other thing about them is they’re ultra transparent.
Every single one of their software tools that they use are all Open Source. They anonymize, and then anybody who wants to can go look at their data and reproduce their results. In fact, they even have Python notebooks where if you don’t trust something that they publish in a paper you can go run it yourself down your own Python and see.
So it’s very transparent from that point of view.
The other company that I would call out is a newer company called Thryve in Santa Clara. They’re focusing on personalized probiotics, but the CEO Richard Lin is an example of the kind of person I like to see running one of these companies because he cares a lot.
He’s been trying to solve some of his own issues, and so he founded a company, essentially, to go and help resolve that. So he cares a lot and he’s especially focused on actionable results. So I like them.
There are lots of other labs; I won’t go into all the names, I haven’t tried a lot of them.
One that I will bring up though is a company called Gencove that focuses mostly on genomics. So they’ll take a mouth sample. But what’s cool about them is that they’ll run their mouth sample, the swab that you give from your mouth, you get the DNA results just like with 23andMe; it’s very comparable to 23andMe. But they also give you the microbiome breakdown.
So there’s that company. And there are lots of other companies that are doing 16S in one form or another.
(0:48:20) [Damien Blenkinsopp]: So that’s very similar to the Atlas Biomed guys, who actually came from Russia. So they were doing studies in Russia, and now they’re in the UK as well, so they got the two populations. So they’ve combined in their interface the DNA and the microbiome.
So it’s quite interesting. I would say they’ve got a lot of recommendations. We’ll get into this in a little bit but they got a lot of recommendations in there, and study references and stuff like that. It’s quite interesting; they’re quite strong on the recommendations from the data.
[Richard Sprague]: Interesting. Do they, what kind of sample do you give them? Is it a mouth swab, or both, or blood? What do you give them?
[Damien Blenkinsopp]: Sorry, this is for the gut, right?
[Richard Sprague]: So it’s just gut, okay.
[Damien Blenkinsopp]: Oh, for the DNA it’s saliva, you’re correct.
[Richard Sprague]: Yes.
[Damien Blenkinsopp]: And then for the gut it’s the usual poop thing.
[Richard Sprague]: Yes, okay.
[Damien Blenkinsopp]: So you do the test and the same time. Or you can send the DNA whenever you wanted.
[Richard Sprague]: So they’re two separate samples?
[Damien Blenkinsopp]: Yeah, that’s right.
They’re trying to combine to get more information, to see correlations, things like that.
[Richard Sprague]: That’s real interesting.
[Damien Blenkinsopp]: And their plan is, I think this will get more interesting. I went to see them last week and so I was talking to them a lot.
And basically their plan is now to get into blood tests as well. And to bring this kind of information to clinicians, where you combine DNA microbiome and blood tests results, metabolomics. And some of the standard stuff as well, like information whatever it is that doctors have been using for a long time. And you can give a bit more context.
So they haven’t figured how they’re going to do that, but the idea is to provide more context around these blood tests to try and make the links and stuff like that to provide a better tool, basically, for looking at patients.
And I think if it’s done that way, led by blood tests which have been used for a very long time in diagnostics anyway, and you add information and context with the DNA and the microbiome, then that actually sound quite useful.
(0:50:11) [Richard Sprague]:That’s right. There’s another company in the US called Arivale, based here in Seattle, and they are now available in the West Coast, California and here. They might be nationwide at this point. But it’s a very similar kind of thing.
I think it’s 1000 dollars, for a one year program. They do a 30x genomic sequence. They test your microbiome, they do your blood test, and there are a couple of other things. They give you a FitBit and measure activity.
And then they assign you a personal nutritionist and you have once a month meetings with them, and you can ask them email questions, and that sort of stuff. And they work with you on whatever issue you want.
And I think that is the direction that I think if you’re seriously trying to solve a problem, that’s what you should be doing. Because it’s this holistic look at the blood results, the microbiome, the genetics and all that stuff together.
[Damien Blenkinsopp]: And consultation. And experts who actually help you work through it.
Because right now, frankly, a lot of these services had to start a consumer facing in order to get the volume of data and build up their databases, right? Because that was the only way that you were going to get enough data to be able to start seeing patterns and start getting past this first hurdle.
And I think it was always sold like that anyway; this is informational, it’s not diagnostic, it’s not supposed to be used like that. That’s really the idea.
[Richard Sprague]: Yup.
(0:51:26) [Damien Blenkinsopp]: Okay. So that’s 16S, and like I said Atlas Biomed that was a 16S as well. And then we have the metagenomic shotgun ones, which I was quite excited about.
I spoke to Eran Segal and Lihi Segal in a previous episode about their work, and that resulted in Day Two. So I was kind of looking forward to that, because it was the first shotgun service to come out that was a reasonable cost. I think at that time it was like 200 or 300 dollars.
[Richard Sprague]: Yeah, that’s right.
[Damien Blenkinsopp]: Yeah. So there’s that one. And you’ve done that as well, and you published a review about it. So what did you think of Day Two?
[Richard Sprague]: I thought Day Two is very cool. You submit the sample, it took a while to get back, but they’re just getting started.
What’s neat about it is Eran Segal, as you mentioned, did a lot of really cool research where they were able to identify, I guess, glucose response levels and it’s dependence on what’s going on in the microbiome. And so by looking just at the microbiome they’re able to tell, oh your insulin levels are likely to respond to what’s in your diet.
And they ran this big machine learning algorithm against all the different kinds of food types. And they had, I think 1000 volunteers and they did a whole bunch of studies.
And now Day Two gives you an app that goes through the food groups and tells you how likely you are to respond, well or poorly, to a particular type of food. It’s very well done.
[Damien Blenkinsopp]: In terms of glucose response, right? It’s just glucose response. So we know that.
It’s been pretty cool. And they had large studies; they had a pretty large population, over 1000 people.
[Richard Sprague]: Yeah. It was, and they’re careful scientists and they published their results.
And kind of the interesting take-away from Eran Segal’s work was that there are some people who, your standard diet advice says you should always eat the whole grain version instead of the white bread version. But there are some people who it’s the exact opposite advice. And this algorithm seems to be pretty good at telling which one you are.
So in my case, for example, with Day Two it’s showing that I should be eating things with more fat in them.
So yeah, there you go Mr. Ketogenic guy. And it was pretty accurate for me. It showed, for example, I’m not lactose intolerant; I can handle dairy and it recommends that I have dairy. And I found most of the suggestions to be reasonable.
The other nice thing about them is they’re not based just on a particular food, but they recognize that food is in context. So having a slice of toast is not the same as having a slice of toast with some butter on it; the way that your body is going to respond is totally different. And they have a lot of recommendations for that.
[Damien Blenkinsopp]: Absolutely.
[Richard Sprague]: So I’m pretty impressed. I’m waiting to see how they do. A lot of the initial research was all done in Israel. So they’re running a study now I guess in the United States. And I think actually when you had them on your podcast I think one thing they mentioned, they’re doing something with Mayo Clinic, I think.
[Damien Blenkinsopp]: Yeah, exactly. Yeah.
[Richard Sprague]: So I’m looking forward to seeing how that turns out in the next couple of years.
[Damien Blenkinsopp]: Yeah, that would be pretty cool when they get more data. Because I think, personally, glucose response is one of the highest impact things you can do relatively simply by changing your diet. Sometimes sleep and other factors as well, but it’s really important.
So going back to this personalized…
[Richard Sprague]: Just one quick thing, did you see the new book that Eran Segal and his co-author put out?
[Damien Blenkinsopp]: No, I didn’t.
[Richard Sprague]: It’s called The Personalized Diet. That’s worth reading. Yeah, that’s worth reading. It’s called The Personalized Diet.
[Damien Blenkinsopp]: Okay, great.
[Richard Sprague]: Go check it. It just came out, and I just read it it’s a wonderful book.
[Damien Blenkinsopp]: Oh, awesome.
[Richard Sprague]: It goes into a lot of… And what’s cool is in the end, he gives suggestions for how you can test yourself using just a cheap glucose meter, and gives suggestions as part of it. It’s kind of cool.
[Damien Blenkinsopp]: Excellent. That sounds a little bit like the Rob Wolf test that was in Wired to Eat.
I put some charts up on that. It’s a standard actually glucose tolerance test to different foods. But you learn a hell of a lot. I don’t know if it’s the same, but it can be done; just a blood meter can tell you a lot of information.
[0:55:22] So I’ve been doing this a lot. One of my other pastimes, currently I’ve been developing a food which uses different fibers because I don’t want it to by glycemic, because I’m not a fan of high glycemic responses. So, similar to the Eran Segal guys.
So I’ve looked at a lot of different fibers and I can tell you that there is definitely a lot of variation between. Because when I go to a company and I ask them for a fiber, there’s many of these. There are a lot of different fibers that are created by companies now in different ways.
Basically fibers are carbohydrate which is resistant to getting broken down in the body. So that’s the way you’ve got to look at it. So there’s a potential high glycemic response from a fiber because your biome may be able to digest it and turn it into glucose, whereas someone else’s maybe not. And it’s going to pass through them and they get no glycemic response.
So I’ve had quite a fun time testing a lot of different fibers and collecting a lot of data on that and seeing the different responses. And I plan to now do that on a population, because I understand that just because I get these particular responses doesn’t mean that everyone’s going to get that response.
But it’s actually tricky with these fibers and everything. There’s a lot of products that state low-carb or whatever, but they often have different fibers in them. And it’s just not that simple, unfortunately.
[Richard Sprague]: That’s very interesting. It would be especially interesting if you could trace it to which microbe is involved.
[Damien Blenkinsopp]: Oh yeah. I know, right.
[Richard Sprague]: There might be a simple little change to the formula, where you add a particular microbe or you add something that that microbe likes to eat and suddenly now that fiber that caused the bad glucose response is suddenly just fine.
[Damien Blenkinsopp]: Yeah, exactly. It’s people like Day Two are going to have the best information because they’ve collected it. I always think about all this whole area and everything, I’ve been thinking about this for quite a whole in terms of us trying to get ahead.
It’s like, who has the data? If you want an answer to something, go find the people with the most quality data it has to be quality data and you’re going to be the closest to the answer at that point. You know, if you can get talking to those guys and what they’re doing with that.
(0:57:29) [Richard Sprague]: Yeah, that’s true. That’s right. So we should also talk about Viome, which is the other metagenomics company. They’re the transcriptomics one that we talked about.
They just came out, and I just got my results back a month or so ago. And again, they give you this big, it’s an app where they’ll give you a big breakdown of the different microbes that you have. Actually, it’s the different, they try to stress that it’s not the microbes themselves it’s the activity of the microbes.
And then they break it down and tell you what kind of foods that you should eat or not. And it’s a pretty impressive list of people backing the company; if you look at their board of advisers it includes people like Ray Kurzweil and Aubrey de Grey, the Life Extension guy, and the bulletproof empire, Dave Asprey is a big fan them. And you’ll see a lot of, Ben Greenfield Fitness, etc.
[Damien Blenkinsopp]: They’ve got their name out in the media more than most companies quicker.
[Richard Sprague]: Yeah, that’s right.
And they’re founder, Naveen Jain, one of the things I respect about him is he really genuinely believes in it himself. So he’s out there himself personally pitching the product, and he’ll talk about his own results. He’s got a private Facebook group where they talk about it, and he’s one of the active participants answering questions about it. So they’re very serious, and they’re hiring a lot of people.
They claim that they’re based on some lab science that was developed out of the Los Almos lab in New Mexico over many years. I’ve had a hard time figuring out from a scientific point of view exactly how they’re doing the work.
One of the things they, if you go to their website they say specifically that they’re not going to release the raw data. So it’s a little hard to tell exactly what’s going on, and how they’re coming up with the recommendations.
And it’s something that I hope that they’ll be a little bit more transparent about.
[Damien Blenkinsopp]: Yeah, and this is something, you know we wanted to talk about, is basically if you’re thinking about doing some labs what kind of things do you want to take into account.
(0:59:24) Let’s talk a little bit about what we’ve actually run. Like what labs have we both used? Because I don’t know you Richard like, what labs have you run over the last, is it four years?
[Richard Sprague]: Well, okay. So, I’m a little crazy. I’ve done well over 500 samples from uBiome, another several dozen from other different labs. Probably all told I’m up at close to say 600 samples.
[Damien Blenkinsopp]: And at uBiome you were doing daily ones, right?
[Richard Sprague]: That’s right. Yes. So I had daily samples for more than a year.
[Damien Blenkinsopp]: Which means you were pooping every day. At least once.
[Richard Sprague]: Yeah, that’s right.
Well actually I should say, I should be more precise. No, not every single day. That’s right. There are a couple of gaps in it, but generally speaking I had near daily samples for more than a year. And then I have other fairly regular samples going back through to 2014.
What’s also cool about it is I tracked all of the food that I ate the whole time, and my exercise and my sleep and that sort of stuff. So I’m able to run all these cool correlations to figure out what I learned. So that’s really cool.
I’ve done also Viome testing, Day Two, Thrive, I mentioned Gencove. Let’s see, who else. I’ve not done any of the culturing tests. But what’s also cool is I’ve done a lot of these side-by-side just to see, to cross-compare them among themselves, which has been interesting.
[Damien Blenkinsopp]: A lot of these labs have interfaces where you have to access the data. So I can’t do it for all of them, but I’ll put up samples of any that I’ve done that are basically PDFs or something that you can actually see.
[Richard Sprague]: Yeah, I’m happy to show mine as well.
[Damien Blenkinsopp]: Yeah, so we’ll combine our things to try and give you a picture of what most of these look like. Can’t be all of them just because some don’t actually deliver the information in that approach, but it should give you a good idea of what all these things look like, and the kind of microbes they’re looking at and stuff like that.
From my side, I started with uBiome when they launched and that’s when Richard also go into it, I believe. And they were one of these Kickstarter campaigns, or that was Indiegogo, because…
[Richard Sprague]: Indiegogo, yeah.
[Damien Blenkinsopp]: Kickstarter and all that kind of stuff…
[Richard Sprague]: Back in 2013.
[Damien Blenkinsopp]: Yeah. This is kind of amazing that it was already that long ago.
So I’ve just done seven uBiome tests. Quite a bunch of those were the five I don’t know if you’re doing the five…
[Richard Sprague]: That’s the five sites, yeah. I’ve done it, it’s gut, mouth, skin, nose, genitals. I’ve done them all.
[Damien Blenkinsopp]: Yeah, I’ve done semen as well, because I was curious. [Laughter] I was like playing around with different stuff. Which they don’t normally do, and they haven’t got a lot of benchmark data on that.
So the standard ones that you said are the mouth, the genitals, and the skin. And they did teeth as well, actually. They did the dental one.
[Richard Sprague]: That’s right, yup.
[Damien Blenkinsopp]: Yeah, so they have quite a bit of data on those.
[Richard Sprague]: Yeah, and we could talk forever about some of the things that I’ve learned from all of my studies. And I’ll give you a link to my, I’ve been writing some of my results up. But don’t forget, the microbiome is more than just the gut and you can learn a lot of things from skin and from mouth and nose as well.
[Damien Blenkinsopp]: Right, exactly. And there’s actually a little hack, we’ll talk about some hacks we’ve done on things that have actually potentially done something in a little bit.
[Richard Sprague]: Yeah.
[Damien Blenkinsopp]: So the other ones I’ve done is the Viome one as well, Day Two, so both of us have done that. I’ve done the Atlas Biomed one, because I’m based in the UK. And I’ve done quite a few of those culture and PCR based tests, so it’s a little bit different there.
(1:02:44) Alright, let’s just dive in to see what kind of things we found from this. First of all, what can we say about comparison of data? We were talking about how they’re not necessarily comparable.
[Richard Sprague]:Yeah. That’s an interesting thing. So I have done comparing my 16S results with both Viome and Day Two, and I find that at the high level, they’re actually fairly different.
I shouldn’t say, you know it’s sort of like you can see the chart here. For example, in Day Two it says that my furmicidies level is about 50 percent. When I tested it on uBiome, one of my uBiome tests shows it’s like 59 percent. My bactorides in Day Two is like 45 percent, uBiome tested it out as more like 30 percent.
There are, that sounds like a fairly significant difference, but if you’ve seen a lot of samples you realize that it’s probably not as significant as it might sound, because there’s a lot of variability in day-to-day anyway.
The one thing that I did notice was that, however, the ordering, in other words which was the most dominant, the second dominant, etc, was pretty consistent. Which is nice to know. That means at least at the biome level you can kind of trust that if it says that you’ve got higher furmicidues than bactroidides then maybe you really do.
The other part is that if it says that you’ve got verrucomicrobia, which is the phylum that includes akkermansia, which is an important one for eating the musilin level and is considered important for health. If Day Two shows that you have it, it’s likely that uBiome will show that you have it as well. Which it’s nice to see a little bit of consistency there.
[Damien Blenkinsopp]: Alright we were talking about this a little bit earlier, because I was comparing all the species that I’ve picked up in different ones. And, you know, obviously they don’t correlate all the time.
So Richard was saying that probably the way to look at it is that if it turns up in two tests, and it’s not in one test, then it could be just that it’s likely it’s there. And it might be worth doing a PCR or whatever, but it’s likely it’s there. And it’s the bioinformatics library of the other one maybe doesn’t include that species, right? They haven’t got the references in their database or something.
But that’s kind of like a starting assumption you can start with in your exploration to try and nail it down, whether it’s there or not.
[Richard Sprague]: Yeah, that’s right. It could be and the other thing, again I would emphasize look at presence versus absence, and be a little bit less concerned about the abundance, and that’s going to vary a lot.
[Damien Blenkinsopp]: Well that’s, on your Viome you’ve got this spirochaete of…
[Richard Sprague]: Yeah, the Viome one is interesting. And I don’t know how to interpret that, because it shows that I have 79 percent spirochaete…
Damien Blenkinsopp]: It’s off the charts compared to the others, yeah.
[Richard Sprague]: It’s off the charts, yeah. And now what they’ll say is that that’s the one that we’re after, those are the microbes that are active.
[Damien Blenkinsopp]: What level is that? Is that the family or the genus?
[Richard Sprague]: It says, my test result says 80 percent spirochaetes at the phylum level, and then it shows at the genus level, the genus spirochaete is 46 percent.
[Damien Blenkinsopp]: So it’s missing one.
[Richard Sprague]: Yeah, there’s just something that doesn’t add up about it, and I don’t really understand how to interpret the results. And I’ve asked them and
[Damien Blenkinsopp]: Right. It sounds like their library isn’t quite there yet, and maybe there is…
For people who don’t know at home, spirochaetes get a bad rep because Lyme Borrelia, which is of course quite a bit of a problem for some people, is a spirochaete. That’s the family it’s in.
So when people see spirochaetes, typically, and when they’re talking about them they’re talking about pathogens. So when you see it in your samples and I’ve seen it in my Ubiome as well. It’s something, I actually did a little project on it, which I’ll, in the show notes we’ll put up anything we talk about, all that usual stuff.
But yeah, I bet you were interested when such a high amount of spirochaetes turned up, and you were like woah okay, but what kind of species is there.
[Richard Sprague]: Yeah, and the results show it broken down by phylum, genus and species. And what was odd is that at the phyulum level it said 80 percent spirochaetes, at the genus level it said only 46 percent, and there were no spirochaetes at the species level.
And the genus level, all of the different genera added up to, I think it was something like 90 percent. In other words, so they think they identified all the genera that were in there, but it didn’t add up. So I’m not sure exactly how that works.
[Damien Blenkinsopp]: Yeah. So I had a little problem as well. When I got my results, I had 30 bacteria in the total, which was showing up, which I felt was relatively low. And so I talked to them a bit about that, and at the time they felt that was correct.
That was when Viome first came out sometime last year. I got my results relatively early. So things may have moved on since then. I would expect as they’re working on the databases and all that kind of stuff that I’ll have more. And I think I haven’t counted them recently, but I need to count them up again but I think I now have more that have turned up.
[Richard Sprague]: Yeah, and they’re pretty clear about, they’re selling a subscription. So right now it’s like 400 dollars a year, or something like that, and so they claim it’s a subscription because they keep updating your results as they learn more information.
So, anyway, so I don’t know how to interpret that.
The other part about it, Viome, like Day Two, has a list of foods that you should eat or not eat. And what I found was there was some consistency between the Day Two algorithm and the Viome algorithm.
For example, both agreed that I can handle dairy products, lactose. Both agreed that I should stay away from grains, although Viome thought that whole grains were okay in a lot of cases. And then there were just some odd ones, like for example Viome says that I shouldn’t eat pork.
[Damien Blenkinsopp]: I think I may have had that too. I had some quite odd things in there.
The issue I had with it was that there’s no reasoning. For the Viome we don’t really know what they’re looking at and why they’re making these decisions. We discussed Day Two, basically we know what it’s based on. It’s based on the glycemic response.
[Richard Sprague]: And there’s an academic paper where they showed the reasoning behind it, and you can, all the caveats that you would see, normally, in any kind of academic study, but at least you kind of know what direction they’re coming from
[Damien Blenkinsopp]: And they’re very focused just on the glycemic response. So you know where that recommendations coming from and they give the A, B, C, D grades.
I would have loved if they showed the average glucose response for someone with mine. That’s what, I actually sent in a support email or something like that in to them for that, because I would be like wow that would be much cooler, rather than these A, B, C, D categories.
[Richard Sprague]: You know they changed it recently, right? They’ve changed it; now it’s not A, B, C, D it’s, they give you a number from 1-10 I think now.
[Damien Blenkinsopp]: Okay. So that’s a bit better, that sounds better. Yeah, that’s good.
Alright, cool. But the problem with Viome is you have no rationale, no methodoloy, and it says you shouldn’t eat something that you love. I think it told me I shouldn’t eat chocolate. So, it’s like, you know I kind of like chocolate and I don’t have any reason.
[Richard Sprague]: Give me a reason, yeah. Give me some kind of…
[Damien Blenkinsopp]: Give me a reason, give me a study. I need something to give up chocolate, you’ve got to give me… Because I don’t even know, maybe you think I’m allergic to it. I don’t know, I don’t know what you’re trying to get at.
So Atlas Biomed has a lot of recommendations as well in their interface, but what I did like is wherever there’s a recommendation there’s always papers, study papers, left there. And there’s always the reasoning.
And you can argue that with 16S and some of the other limitations they have, maybe they’re pushing the edge in terms of their recommendations, but at least they’re trying to give, you know, a reasoning and structure. And there’s a transparency.
So, with Viome, the thing for me is it’s not transparent, so you can’t, you don’t know what you’re getting, what the output is. So it’s like, how can you do anything with it really. At the moment.
[Richard Sprague]: Yeah, you kind of have to trust their scientists, or whatever the results is of this thing. Yeah.
And the other part of it is, remember also it might say, eat apples. Well, there’s lots of different ways you can eat apples. There’s a Fuji apple that’s different than a this kind of apple, there’s an apple that was just picked versus one that has been sitting in a truck for a while.
There’s lots of different kinds of things. And to just say a blanket statement, eat more apples, is, you know, I don’t find that as scientifically satisfying as it could be.
That’s why I like the Day Two approach more to talk about, well we’re not going to say apple versus not apple; we’re going to say apple with cheese versus a meal made out of apple pie, or something like that.
[Damien Blenkinsopp]: Yeah. I was talking with a guy who runs another bioinformatings company just the other day about this, and basically a lot of people have a religion about food. It’s not like everyone’s really objective about this.
Vegans are vegans, and ketogenic people are ketos I’m guilty of that one. And it tends to be an emotional thing. I try to be more objective and numbers driven, but, you know…
The problem is also, when we’re doing these tests, if you tell me not to eat my favorite vegan food and I’m a vegan, you’ve really got to and the argument is, say, glycemic response, and a lot of vegans don’t care about glycemic response, right? I think.
So if you actually gave us the reasoning, then different types of people with different approaches and thinking towards their eating style will be able to choose. They can be like, but I don’t care about that. I don’t care about glycemic response, or I don’t care about the other factor, or I don’t care about allergies. Or whatever the reasoning is. And at least that would give you a better framework in order to make a decision.
[Richard Sprague]: That’s a good idea, yeah. Have you used Inside Tracker? The blood testing company, Inside Tracker?
[Damien Blenkinsopp]: I haven’t. I know they were on a show a while back.
[Richard Sprague]: That’s another company I have a lot of respect for. It’s not the microbiome, but they have, it’s all about blood testing. And they’ll do exactly that. You can type in, you could say, I’m a vegan. Now give me your suggestions. Or, I’m a carnivore, now give me your suggestions.
And it’ll be tailor-made for you, because they recognize, like you say, that you may have another framework that you’re thinking about. And if your diet suggestions can’t fit in my framework, I may have to either give up my framework, or maybe I’ll give up you.
[Damien Blenkinsopp]: And this is something I’m seeing more in my results. When their recommendations come up and when I’m looking at them, I’m like oh, you know, that doesn’t fit with the ketogenic diet. That’s where I am currently.
[Richard Sprague]: Yeah.
[Damien Blenkinsopp]: So you want me to eat more of that, but I’m just not interested.
[Richard Sprague]: Yeah.
[Damien Blenkinsopp]: So there you go, even if I’m being objective. But if I had more information I might reconsider it a bit more.
[Richard Sprague]: Yeah, exactly.
(1:12:55) [Damien Blenkinsopp]: Okay, so what other kind of interesting stuff have we discovered here?
The other contrast, like I was referring to, I was trying to do earlier, was the Aperiomics, which is a shotgun sequence as well. And I was trying to compare it with the PCR to identify similar things. But that didn’t quite go as well, either.
So I think the shotgun technology, although it’s more detailed than the 16S, it’s going to take time for those databases and bioinformatics pipelines to evolve so that it’s picking up everything.
[Richard Sprague]: Yeah, I think you’re right. And like I said, I think you probably can trust a single lab over time. So if you’re doing A/B testing on a particular kind of intervention, and you follow the same lab both times, you may be able to trust that. But looking at the results from different labs, I just don’t know how useful that is a lot of times.
Especially when you get down to the species level, or down to something very, very particular. There’s just too many ways that they can be different.
(1:13:51) [Damien Blenkinsopp]: So because I’ve mentioned the ketogenic diet, one interesting thing is that if you look at some of the studies they suggest that if you’re on a ketogenic diet so I’ve been on a ketogenic diet for something like, since 2011, and then really seriously since January 2016. I was actually blood testing and stuff to make sure.
What they say is you should see increased microbes of the genus bacteroides and decreased firmicutes. And if you look at all my early uBiome tests, 2014, 2015, 2016, a lot of the time it’s the opposite.
[Richard Sprague]: Hmm.
[Damien Blenkinsopp]: Yeah, and I’m firmicutes dominant. I remember looking at this when I was first, I was like that doesn’t really sound like me.
And I think this goes back to the papers sometimes, as well. The studies when they’re looking at these things. I’ve got a team working looking at them, ketogenic studies and stuff like that.
When you look at a lot of the ketogenic studies, they have very different diets in them, unfortunately. You know, 40 grams carbs, 5 grams carbs, 50 grams carbs and doing different things. So a lot of things, when you look at these studies, even, you have to kind of look at the details of the studies. What they were actually doing, and then the diet.
So, you know I complained, I think. And I would bet that the reason I’m getting a different result there is because I have a, what I would call, a well-formulated ketogenic diet. Which means that I eat a lot of vegetables and, you know, fibers and things like that.
Because I think the main hypothesis there is that someone on a ketogenic diet is eating less fiber, basically, to feed his gut biome, and therefore you’re seeing that inversion.
[Richard Sprague]: Oh, I see.
[Damien Blenkinsopp]: But I’m not seeing it, so I think its because the type of ketogenic diet I’m running is different to that. So even when you’re looking at some of these studies, you have to be careful to look at the details of them as well, and does it exactly resemble you.
[Richard Sprague]: That’s true, yeah that’s true because not all ketogenic diets are going to affect the microbiome the same way. Yeah, that’s right. And then you get into the whole definition issues, of some people say that this or that is ketogenic and other people would dispute it. Yeah, that’s all tricky.
(1:15:54) Let’s talk about some of the things that we’ve done. In your show notes, I hope we can put some of these images that I’ve put up here, but there’s one in particular I guess if you’re asking me my take-aways. I think people need to recongize that a broad measure, something like diversity, which is something a lot of people care about, it’s real hard to tell what that means. And it’s very hard to just put a single number on the concept of musilin.
We all sort of intuitively understand that having a diverse microbiome is a good thing because you’ll be able to respond better to different challenges that might come up in your environment. But if you have a diversity of pathogens that’s not necessarily a good thing. It sort of depends on what’s in there.
And the other part is, and this is true of generally I find through daily microbiome testing is that there’s a lot of variabilty day-to-day.
So one of the charts that you can look at in here is just showing the diversity that if you tested me on a Monday you would say I have low diversity. In this case I have like 1.8. But if you tested me on Tuesday I was all the way up 2.3. And then if you wanted until the weekend, by Saturday I was at, maybe, it was still hovering around 2.1, but then suddenly on Sunday I plunged to under 1.8 again.
[Damien Blenkinsopp]: So we understand, with these diversity algorithms, right, that they’re running, is that looking at species diversity?
[Richard Sprague]: No it’s looking at the family level, which makes sense because the family level is kind of a good level to look at because you still have a lot of coverage. You’ll get close to 100 percent of all the different things that are there, unlike say genus or species where there are lot of ones that just won’t show up in the 16S.
[Damien Blenkinsopp]: In the 16S, yeah they won’t show up so you wouldn’t be — yeah that’s what I was getting at.
[Richard Sprague]: So they test it at the family level. And there are a couple of different ways to measure, but one way to measure it is, you can think of it as the probability that if I grabbed two things at random, two microbes at random from my gut, the probability they would be the same,
And in the case of if you, for example, if you’re firmicutes dominant and a lot of people would have 70 percent firmicutes, it’s pretty likely that if you grabbed two random microbes that both of them will end up being firmicutes. But it’s very unlikely that two of them would be something else, and that’s the way you measure diversity.
There are a couple other different measurements for diversity, but they all rely on the this idea that in aggregate we’re looking at, like how much information is in this signal. And that’s a little difficult to be able to really pin down.
Now that said, the other thing that I pointed out is that although it’s variable day-to-day, if you look at my picture and we can put this in the show notes too if you look at my diversity across the year, yeah there’s a lot of day-to-day variabilty but there’s a trend. There’s kind of an average there.
And I’ve looked at this with other people as well, and it’s unique to me. So there’s something different, something special about my gut that is different than your gut. And even though there’s a lot of day-to-day variability in how that works, I think there really is something there. There’s some kind of signal, we just have to understand better what that signal is.
[Damien Blenkinsopp]: Right. So you’re saying diversity is interesting but we don’t understand why it oscillates.
[Richard Sprague]: Yeah, and it’s partly because we don’t understand diversity, or know what that really means.
[Damien Blenkinsopp]: Well I think it would be really interesting. You’re saying it works at the family level, and that’s because…
[Richard Sprague]: That’s how we measure it, at the family level usually.
[Damien Blenkinsopp]: Right. So that’s what we’re measuring currently. And it’s not the ideal, right. I mean, ideally, maybe with the shotgun. And I don’t know if there’s studies actually on this. Because I’m assuming that the studies were all done on a 16S for diversity.
[Richard Sprague]: Oh no, people do diversity metrics for any sort of sequencing.
[Damien Blenkinsopp]: Okay. So they’ve done it on shotgun as well, but they still do it at the family level?
[Richard Sprague]: No, just generally speaking, if you want to be able to compare two different samples that were done on 16S, you’ll probably want to compare at the family level.
[Damien Blenkinsopp]: Yeah.
[Richard Sprague]: But there are other ways to measure diversity too that might be useful. Like just counting up the total different number of species that were found in your sample versus my sample. And you might find that you had 150, I had 130. And that’s kind of interesting to know that you have some microbes that I don’t have, and maybe vice versa.
[Damien Blenkinsopp]: Yeah.
[Richard Sprague]: But that’s hard to capture in a single number, and a lot of people, like the Viome test wants to be able to say in one chart, what is your diversity. People sort of care about that.
I’m just, in my experience that’s hard to pin down.
[Damien Blenkinsopp]: Right, and it’s hard to say it’s actionable or you can even say, okay I’m diverse, I’m well. It seems too abstract in terms of a biomarker.
[Richard Sprague]: People who complain about having low diversity, I’d say why don’t you test yourself tomorrow and see. You might like the test results you get better tomorrow, I don’t know.
(1:20:15)The other, just to quickly show you one more of my charts that I think is fun.
So I tested myself doing a probiotic, taking a pill, to see what would happen. And in this chart you’ll see there’s a little red splotch on there that shows there’s about a nine or ten day period that I was taking this pill daily to try to improve my levels of bifidobacterium.
And on this chart you’ll see that it’s hard to see there’s much difference in the level of bifidobacterium, but there’s another huge spike in my bifidobacterium that happened several months before I took that.
[Damien Blenkinsopp]: And we are talking huge, guys. You’ve got to look at the chart.
[Richard Sprague]: It’s totally, totally different. And the fact is that that month of September I happened to be traveling in New Orleans and eating a lot of red beans and rice, which apparently affects my bifidobacterium levels. And that’s kind of the take-away lesson for me is that often the best interventions you’re going to have are going to be some kind of food that you eat.
[Damien Blenkinsopp]: Probiotic.
[Richard Sprague]: Like a prebiotic, yeah. Because I think what’s going on is these microbes all interact with one another. And so just increasing one is sort of like poking on one little thing hoping that that’s going to improve it, but really that’s going to create a cascade effect of a whole bunch of other things.
[Damien Blenkinsopp]: Absolutely.
[Richard Sprague]: And the only way to really improve things is probably holistically.
[Damien Blenkinsopp]: Yeah, yeah, absolutely. It comes back to the whole foods approach and everything, right? That we can’t approximate, we can’t invent a food with our food science because we don’t fully understand what’s in a whole food. Right?
That was one of the concepts out there. And so we should just eat whole foods and then we’re going to get everything that we need. And one day when science has really understood all of the tiny details we can maybe mimic it. But for now it’s probably just not a good bet to be able to do that. So, really interesting.
[Richard Sprague]:Yeah, that’s right.
(1:22:01) [Damien Blenkinsopp]: One of the things I came across in terms of a test was putting kimchi up your nose.
[Richard Sprague]: Yes, I’ve heard about that, yes.
[Damien Blenkinsopp]: Right. Because I think we discussed it before on a past call. So this was something recommended to me by a physician, because I had experienced some sinus headaches.
And people have been experimenting on this we’ll put up the links on the internet and blogs about this approach to reducing the incident, or eliminating, sinus headaches. And basically there’s certain types of kimchi that contain cayenne, which is in all of them today because there’s a lot of different kimchis on the market.
And they have to be unpasteurized. And basically you take, you don’t put kimchi up your nose literally. Thankfully, you take some of the liquid in that, so you put a teaspoon in, pull it out, then you dip your finger in it and you put your fingers up your nose, both nostrils, to get some of that in there.
So you’re snorting the juice, basically. And the idea is you get lactobacillus sakei up there, and that helps to populate the nose if you’re doing that every day. And that helps to counter some of the microbes that are potentially causing the sinus headaches by their overgrowth. So it’s countering their growth, basically.
It did seem to have a positive effect for me, but unfortunately I wasn’t doing any biome test or anything like that at the time, so there’s no data on that. It’s just an idea that someone might want to test. And I’d love to see some biome or something else results on it if you do do it.
[Richard Sprague]: It would be interesting, yeah. So my daughter suffers from sinus headaches now and then, and I told her about what you had suggested. We have this big jar of kimchi still in the refrigerator, but she just wasn’t interested.
[Damien Blenkinsopp]: It’s kind of a weird sensation at first, I have to say.
[Richard Sprague]: It’s like the other advice that I got on the internet was you should simulate, what is it called, the brain burn that you get if you have some very cold ice cream or something. And she doesn’t mind doing that, eating a lot of ice cream when you have a headache. But kimchi up the nose thing was a little bit hard for her to try.
(1:24:06) [Damien Blenkinsopp]: So obviously there’s a lot of probiotics on the market right now. A lot of them, and I think going back to what you were talking about, when you introduced one of those into this environment and we have been talking about that there’s a homeostasis of that environment.
They work together, they feed each other, and you just throw one in there, he’s basically getting thrown into an alien population. Because if you’re adding them, it’s probably because you don’t have them, so it doesn’t really fit in with that environment right now. And that’s my assumption why they’re not growing, not sticking in a lot of results like yours that you’ve seen.
Because it’s probably, he depends on some other guys, some other bacteria. That would be interesting studies. Like, bifidobacterium, everyone knows that these are beneficial, what other species do we need in there to support them, and then concoct basically a probiotic which maybe allows that. And maybe adds prebiotics as well. I mean, that sounds good to me.
[Richard Sprague]: Yeah, and people tried doing that. And I’ve looked at a lot of people who’ve done A/B testing, where they test their microbiome before and after, and I have yet to see convincing evidence that any of them makes any difference. Yeah.
[Damien Blenkinsopp]: Right. And they’re quite expensive, some of them, right now.
[Richard Sprague]:Yeah, that’s right.
Now, that doesn’t mean that it doesn’t work. And there have been studies like BSL-4, I think, is the one that people talk about. They’ve done randomized controlled trials and they show that such-and-such marker is actually improved, or such-and-such disease state is improved after taking the probiotic. I just haven’t seen that demonstrated…
[Damien Blenkinsopp]: In the data. But that’s also like, okay, so maybe it’s something that’s not being picked up in that particular sequence, the bioinformatics pipeline, or whatever. And it will turn up in two years when we’re finally tracking it. That’s the problem with where we are right now; something could be happening and could be beneficial, and we’re just not finding it in the data is all.
[Richard Sprague]: Yeah, who knows. Or it could be that they way they do the testing, these randomized trials, maybe they all drink a glass of orange juice after they take… Who knows.
Yeah but I do think in general, a lot of people ask me after all my testing, What do you think about taking probiotics pills?
And my general, I just have not seen any good evidence that any kind of pill really helps. If you want to make a difference to your microbiome, do something involving food.
[Damien Blenkinsopp]: And a variety. I think a wide variety makes sense. If you’re trying to get diversity, a variety of vegetables which is supposedly a good rule of thumb for micronutrients and other reasons as well it can’t be a bad thing to do.
[Richard Sprague]: Yeah. You can have, you can put up a link to the, I’ve got a medium place where, medium.com, where I posted a bunch of my microbiome experiments. But a few of the things I’ve tried are like, kombucha, soy lint, makes a difference in the microbiome.
[Damien Blenkinsopp]: Oh right, that’s an interesting one. Yeah, so the whole, whole…what do they call it. Nutritionally complete food.
[Richard Sprague]: Yes, right.
[Damien Blenkinsopp]: Yeah. There’s like 60 companies that have started those now. I didn’t realize until I looked into it the other day. Didn’t you do a colonic at one point? Was that you?
[Richard Sprague]: I did, I did that as well. Again, my take-away was that I was hoping that there would be some ability to make a major change afterwards, by feeding myself the right kind of things. But it just bounced right back to the way it was. Two weeks later I was right exactly where I was before.
[Damien Blenkinsopp]: But that’s actually, that was good feedback for me because I spoke to one physician who’s been working in environmental medicine for a very long time about something that I had.
And he suggested six colonics within two weeks. And he didn’t know why, but he’d been doing it for 30 years. And he said, I don’t understand completely the mechanism, but it really helps with this specific thing.
So, I did it. But I was concerned about my biome, obviously, doing that and colonics and stuff. So when I heard your story I was like, okay.
[Richard Sprague]: Yeah, and who knows. I’m just one guy, so.
[Damien Blenkinsopp]: Right. N=1.
[Richard Sprague]: The other thing that people should realize based on my experiments that I don’t have an appendix. It was removed when I was five years old. And the appendix is known to include, that’s where the bacteria gets stored when you…
[Damien Blenkinsopp]: It gets stored. Yeah.
[Richard Sprague]: Yeah. So who knows what’s going on in my gut.
[Damien Blenkinsopp]: But that’s a good test though, because then you don’t have that storage device, basically.
[Richard Sprague]: You’d think, yeah, but who knows.
[Damien Blenkinsopp]: Yeah but that is a pretty important N=1 difference there.
[Richard Sprague]: But nevertheless, for me at least, everything just seemed to bounce back. And I’ve found that my microbiome is pretty resilient to just about any kind of change.
[Damien Blenkinsopp]: Yeah, hard to change.
[Richard Sprague]: Yeah, that’s kind of the bottom line.
(1:28:22) [Damien Blenkinsopp]: Alright, so we’ve dived through some of our own personal experiences there, trying to change it. And as you’ve kind of heard it’s not easy to change your microbiome, it seems. But it doesn’t mean it’s not worth experimenting with.
(1:28:34) So the thing I’d thought we’d do now is kind of take a step back and look at the big picture of all of these labs and everything. To see where they are and what kind of, you know, thoughts we have about using them, I guess, right now. What’s valuable to you, you the guys at home, to be doing with these right now and potentially in the future.
Richard, what are your overall thoughts?
[Richard Sprague]: Well, so, it’s hard to beat the price of 16S. And it is something that’s also pretty easy to do; you don’t have to poop in a box, you don’t have to put tablespoons, laying on the floor kind of thing like this. It’s relatively easy to do. And for that reason alone, I think it’s worth doing a 16S test. Do a couple over time, or if you’re trying to check out the effect that it has on one particular thing, it’s cheap and easy.
[Damien Blenkinsopp]: If I can just jump in there, I think that’s interesting also because of what we’ve said about the bioinformatics pipelines and the databases will be evolving and getting better over time. And that sample is part of your history, which could be useful if and this is actually Jessica, she came on the show way, way, way back and she suggested it was good.
Say you get sick in the future, it could be gut related, and you have that sample. As the bioinformatics and the database evolves, you could then look back at that and be able to see what the difference is. And you would be able to formulate some kind of plan to try and get back there, at least.
So just for that reason, for this historic storing your sample if you ever need it in the future, it’s a reasonable idea.
[Richard Sprague]: Yup. I think that’s something everybody should do. And we talked about the other tests.
I told you about Day Two, I like the science behind them. It’s like 300 something dollars, I guess. Little bit expensive, I think, but a lot of people would find it would probably be useful for you if you were looking at a particular condition, particularly any of the metabolic diseases like diabetes, I would think that you would want to do this.
Because it’s going to tell you based on these peer-reviewed studies, it’s going to tell you something about your glucose response to different kinds of foods.
[Damien Blenkinsopp]: Yeah. If you’re overweight, if you’re really overweight, it’s probably interesting.
[Richard Sprague]: Yeah.
[Damien Blenkinsopp]: Because it might just pick out one of those foods that is your main go-to every day.
What I like to think about these kinds of tests, because we’re saying it’s not 100 percent, but it’s a good broad picture. And if some of these foods that you’re eating every day come up as red in their algorithm, you can then go and test them properly. And you’ve saved a lot of time and effort because it gave you that broad look at all of the foods.
And it gave you some way of basically strategically focusing on like five different foods that you’re eating a lot and turned up red there. And then you could do a proper glucose test with a meter on each of those. Whereas obviously you couldn’t do that on the thousands of different foods you’re eating, or hundreds, that you’re eating each week.
[Richard Sprague]: Yeah, that’s right. And like the example I give is I have always eaten a lot of bananas. Now, I’m aware of the carbs, and the sweetness, and everything else. Bananas I always thought were nutritious; it’s a fruit, it’s healthy and it’s easy to eat.
But both my tests Day Two and Viome results came back saying that I should avoid bananas. Which I thought that’s kind of interesting evidence. And so that’s the kind of thing I probably wouldn’t have thought about.
[Damien Blenkinsopp]: Have you, my first question, have you tested your glucose response to that? I’d really like to see.
[Richard Sprague]: I mean I test my glucose response, but I haven’t noticed any major differences. I have not tested my glucose response after eating a banana, I should do that. It would be interesting.
[Damien Blenkinsopp]: Right, that’s what I would love to see, to marry that up. Yeah.
[Richard Sprague]: Yeah that’s a good idea, I’ll try that.
[Damien Blenkinsopp]: Please do, and we’ll chat about it later.
[Richard Sprague]: Yeah. And I guess those are the big commercially available ones in the US. You mentioned Atlas Biomed and Aperiomics is that what you said?
(1:32:11) [Damien Blenkinsopp]: So Aperiomics is designed, their whole thing is focusing on pathogens. They mostly work with physicians , and they mostly get people who have strange illnesses and haven’t been able to figure anything out. I mean, she’s got some interesting stories.
I’ll tell you, because I’ve been talking to the girl that runs the lab. And I figure I’m going to use it a little bit more, because it appears like a lot of us and I’ve been talking with other scientists about this a lot of us carry a bunch of pathogens around with us all the time.
Depending on where your immune system is and everything else, you could be fine. But that doesn’t actually mean you want to harbor these things for the rest of your life. Because they do see some correlation later in life to certain neurological diseases and stuff to some of these pathogens.
And so I think it’s a preemptive. Because I’m a bit anal, I’m quite interested in that to screen for certain things that I might decide to try and remove for the long-term benefits of removing those things.
So I’m actually going to run a blood sample through her as well. But she’s got some interesting stories. Like she had some patient come in with some sample and they couldn’t figure out what the problem was. And it was a species of leprosy.
[Richard Sprague]: Ooo, okay.
[Damien Blenkinsopp]: Yeah. And apparently this specific one isn’t supposed to be around anymore. So they’re picking up stuff that is kind of presumed dead or gone in the past.
So I think her lab will be interesting. I’m not sure how fast she’s accumulating data. But if anyone’s got something, some really strange medical condition out there it might be an option to just try and get some ideas on the table.
[Richard Sprague]: Yeah, I think that’s a good point that especially for people who have some kind of misdiagnosed chronic condition, where your doctor and maybe doctors, and you’ve consulted lots of people and they don’t know what’s going on. And they’re just, can’t figure it out.
I do think that any of these tests is going to be valuable as an additional data point. Now whether it’s going to produce something actionable for you or not, I don’t know. But I’m really glad that we have the technology available for us to better ourselves.
[Damien Blenkinsopp]: Yeah, it’s exciting. It’s starting to give us ways to try and decipher these mysteries. Or at least get us closer to the results more quickly. And often it’s kind of leads.
Obviously it’s not, that’s why they’re not being used by physicians that much is because they can’t give you a diagnosis. But they can give you leads and patterns, and eventually someone can figure something out from that.
[Richard Sprague]: Yeah, yeah. And like my example of bananas, I think that a lot of times just doing a different test like this will maybe point out something that you had not been focusing on; you had sort of taken it for granted that this was just the way things are.
This is the way that I live. And sometimes they’ll kind of shake you up a little bit and say, well wait a minute. Have you tried rethinking this previous assumption? And I think that’s valuable too.
(1:34:52) [Damien Blenkinsopp]: So what did you think on the 16S versus shotgun? I mean, they’re not that far apart now in terms of price.
[Richard Sprague]: Yeah, I mean again. Well, I mean.
[Damien Blenkinsopp]: It depends on your budget, right?
[Richard Sprague]: Depends on your budget, yeah. And I know a lot of people who would say 400 dollars, or 300 dollars, is a lot of money to spend on something that’s not quite that well understood. And I understand that argument. I think that if you can, I think it’s definitely worth it. I think you’re getting some new insights that you wouldn’t have had otherwise.
We talked about the question that we have about the transparency of the results of Viome and where they got their…
[Damien Blenkinsopp]: I think transparency is key because, it’s also, I think it’s a little bit about the ethos of the company. Like the ones that are already transparent, you can see, as we were saying, these samples they have them, they’re going to evolve over time. So it’s going to become more valuable provided that it’s transparent.
[Richard Sprague]: That’s right, yeah. Yeah. I do want to know why it is that you gave that recommendation, and then I want to know and trust that if someday you discover new science that makes you retract your recommendation then I’m going to hear about it. And you’re going to be honest and up-front about it.
[Damien Blenkinsopp]: Right.
[Richard Sprague]: Because here’s the thing about science. Real scientists, they want to be proven wrong. They’re constantly working, it’s why their…
[Damien Blenkinsopp]: Exactly. The search for truth. Yeah, exactly.
[Richard Sprague]: Exactly. And I always get a little suspicious when I’m talking to one of these companies where they act like, What do you mean, are you questioning my science? Are you questioning my results?
You know what, yeah. They should be glad for that.
[Damien Blenkinsopp]: Well they don’t give you full access to the data. If you don’t give me my raw data, I get nervous.
[Richard Sprague]: That’s a red flag right there, yeah.
[Damien Blenkinsopp]: Yeah. So, Richard just brought that up. We’ve got a little table here we’re going to throw up. He was like, oh yeah raw data, and I was like damn I forgot that one.
(1:36:30) When you can, raw data is going to be really helpful. And it just proves that they’re transparent as well. I think that’s a really important thing when you’re going for one of these services, to ask about.
[Richard Sprague]: Yeah.
[Damien Blenkinsopp]: And I think most of them are going to provide that. We spoke a bit about, some of them haven’t done it quite yet but they say they’re going to do it soon.
[Richard Sprague]: Yeah. I look at it as a reputable lab will be happy to give you the data because their real value that they had is in the interpretation side. And they have access to additional, maybe proprietary data or insights that you don’t have.
Which is fine, that’s where they’re going to be differentiating themselves. But the raw data itself, it’s just data. It just comes right out of, it’s your data; it’s about your health. You should be able to look at it. That’s my attitude.
Plus, in the case of something like uBiome, one of the reasons I’m very, very excited about uBiome’s raw data is that we’re able to go and take that data and do things with it that uBiome just doesn’t have the time or the, maybe just the bandwidth to go and pursue. And so a lot of these charts that uBiome sent me, I did that because I had access to the raw data. I wouldn’t have been able to do that otherwise.
[Damien Blenkinsopp]: Yeah. I actually had, anyway. That’s a long story. I had a little project to identify a species which I thought would be useful to the 16S. Basically like a strategic screen for pathogens using some tools. So I actually got that sent to uBiome, and they were like, ìThis is really interesting, but we have a lot of other projects that are taking up all our time right now.î
So there’s a lot of stuff these technologies could be used for in the future. And I think that’s one of them. A very cheap method for some doctor to get a strategic screen, and then for pathogens, for a list of pathogens. And if something comes up, you then do the PCR, which is more expensive. But you’ve done it really cheaply. So I think that’s going to be, hopefully, a really interesting application in the future.
[Richard Sprague]: Yeah, that’s right.
(1:38:23) [Damien Blenkinsopp]: What other things do you think might be cool in the future? Or what applications do you think these are going to turn out to be pretty useful for? Or, what do you think you would use it for today, if you’re going to use it for something?
[Richard Sprague]: Like I said, I think that most people talk about gut microbiome, but there’s a lot of interesting things you can learn in the other microbiomes as well. And I think we’re going to see a lot, in the future I think we’re going to see more emphasis on, say the mouth and the skin. And there’s just these very intriguing associations.
For example, one of the things about Alzheimer’s disease, one of the early symptoms of Alzheimer’s disease is a lack of smell. And there is some evidence that the nasal microbiomes of people with Alzheimer’s are different than those who are not. And could it be that there’s a microbe that just sits in the nose for years and years, decades and decades, and finally migrates into the brain and that’s what triggers the disease?
And we’re going to find all kinds of associations like that.
[Damien Blenkinsopp]: Yeah. And I think it’s often going to be multi-factorial as well. And that’s why data from all of these places is going to be so invaluable, because we’re going to be like, oh look, when you get these 20 factors together.
I mean this is why we haven’t been able to figure this stuff out yet because we focus on one factor, and we just can’t see the big picture. Which is way more complicated.
(1:39:39) [Richard Sprague]: And talking about AI is becoming kind of a buzzword, but I do think that the ability to be able to go and look at all these different tests all holistically and be able to look at all this different data and then see patterns, that is one thing that AI is good for.
[Damien Blenkinsopp]: Yeah.
[Richard Sprague]: And we may be surprising ourselves in the kinds of insights that are possible.
Damien Blenkinsopp]: I know, right. It’s going to be really interesting what comes out. And some people are going to get really annoyed by some of the stuff AI brings out. It’s going to trash some stuff we’ve been doing for a long time.
[Richard Sprague]: It probably will.
(1:40:05) [Damien Blenkinsopp]: One cool thing that’s actually going on is, and Richard eluded to this with another company before, but Day Two, what’s interesting about these companies is they’re evolving pretty quickly as well.
So Day Two when I did it, was just a test. But now when you buy it, you actually get a nutritionist consultation. So they’re embedding that with it.
[Richard Sprague]: Yeah, that’s right.
[Damien Blenkinsopp]: Did you get that when you signed up for it?
[Richard Sprague]: I just, I never talked to the person. I probably should at some point.
[Damien Blenkinsopp]: You, follow up. I’d love to know what they talk about.
[Richard Sprague]: Yeah, well the frustration I was having is because I want to know a lot more technical details about stuff, and they usually don’t know the answer.
[Damien Blenkinsopp]: Well, you find out. You never know, you might hit the jackpot.
[Richard Sprague]: Yeah. In the case of both Day Two and Viome I was very impressed that they reached out to me. I got a call out of the blue from someone from Day Two, and they just said, We wanted to talk to you about your test and what you thought about it.
[Damien Blenkinsopp]: Wow.
[Richard Sprague]: It was like, how did you get my phone number? And they said, ìWell you put it down when you registered for the product, and that’s what we’re calling it for, because we wanted to know what you really think.î And I chatted at length with somebody and told her exactly what I thought about the product.
So I’m encouraged that they are going out of their way to do this. Similar with Viome, I know; they are calling people up and saying…
[Damien Blenkinsopp]: Are you saying uBiome, or is there another company?
[Richard Sprague]: No, Viome.
[Damien Blenkinsopp]: Oh, Viome. Okay, got ya.
[Richard Sprague]: They’ve been very proactive about making sure that people send their samples in, and find out why it is that you’re not sending the samples. So I’m encouraged that the whole industry is undergoing this kind of push to be more customer centric. And maybe really trying to solve people’s problems as opposed to just a fast way to make money.
[Damien Blenkinsopp]: Yeah, exactly. Solving results, giving people results is where it’s at.
[Richard Sprague]: Yeah.
(1:41:44) [Damien Blenkinsopp]: Okay, so what do we think are the things that have to be improved? I mean, we kind of touched on this already, but to get to something that’s going to be far more valuable, like all of these services, so that we’d be saying everyone should be getting these done and really using them.
What has to happen with the technology? What has to happen, and do we have any kind of reasonable timelines?
[Richard Sprague]: So there’s kind of a movement. A lot of these companies are trying to add better access to the literature.
So for example Thrive now, they’re proud of the fact that they did some kind of machine learning thing, where they went through all the literature and looked at all the references to different microbes, and they’re going to tell you this and that about it. So there’s some activity around that. I think that’s helpful.
I think it’s nice to be able to have some way other than just googling the name of a microbe to find out what it is. If we can get more into, more vetting of the literature that would be good. Even if you spend any time with this area you’ll notice that there are thousands of new articles coming out, new journal articles, new peer-reviewed journal articles coming out every day.
You can’t keep up with them all. And a lot of them are self-contradictory. It’s just very hard to tell.
So if maybe there was a little bit more emphasis on curating the results a little bit better, that might be useful.
[Damien Blenkinsopp]: Standardization. Somehow.
[Richard Sprague]: Right. The way the labs really report the results, the way that they publish the results. That kind of standardization I think would be great.
[Damien Blenkinsopp]: So I was talking with a bioinformatist who’s working in the nutrition area. He’s got one of these apps similar to, that tracks food. Food intake and all of that. And he was telling me that the databases that all of the companies with these apps, all of them, are using are really low quality.
So then it’s a very similar instance, and I’m sure it’s similar in most of these areas where the quality of data is actually very poor. And we’re just talking very basics here, like the macro content of a food, which is being put in their database. Then you take the photo, or you enter, you pick it from their library, and you think you’re getting that macro content but you’re not because the data is quite bad.
So they’ve personally just been building a very low volume database. So it has less in it, but it’s high quality. And they’re thinking about just throwing it out there as Open Source to try and bring the industry up a bit. To try and get people using that and building on it. And improving on it.
But I think what’s happened is a lot of people have been conscious that their databases aren’t broad enough, or don’t have enough volume in it. So it can be very frustrating for customers and all of this stuff. So they’ve chosen other approaches like just get customers to add the information in, or whatever. And these are low-quality approaches, and then you end up with a lot of garbage, unfortunately.
So, this is a very important topic for quantification in general, and getting actionable information out of it at the end of the day.
[Richard Sprague]: Yeah, and everybody kind of wishes there was Wikipedia of knowledge about the microbes and about the food benefits and all that kind of stuff where anybody can go and add their results. I guess that’s Wikipedia.
[Damien Blenkinsopp]: There is value in crowd sourcing, but it’s those processes and things that you have to put in so that you get a volume, but then it’s filtered, and filtered, and filtered. So that you maximize the benefits of building volume through crowd, but at the same time there’s that mechanism to ensure that quality eventually ends up there.
[Richard Sprague]: Yeah, and it works for things like, MyFitnessPal has any kind of food you can possibly imagine in any culture, any language, anything and they’ve got it in there. Because they’ve got this crowd sourced thing figured out to a science.
And in fact they were telling me that when Nabisco put out some new packaged good, they had the calorie information in their database before Nabisco.
[Damien Blenkinsopp]: That’s ridiculous.
[Richard Sprague]: It’s like somebody just immediately.
Yeah, but you know it’s of unclear quality. And in a lot of cases, particularly with foods, and with the microbiome, like we were talking about apples; there’s a lot of difference between what was tested in a lab somewhere and what you’re actually putting on your plate.
[Damien Blenkinsopp]: Yeah, and I can tell you, because I’ve been digging into food science and stuff for one of my companies, and when you see an ingredient on a label, there are 20, 30, 40, 50, 60 different versions of that that would fit into that name. And they have quite different properties in some instances.
We’re picking three different ones, and we’d go through ten of them until we get to one that does what we want to. So there can be a lot of variation on this. So when you’ve got these ingredients and they’re using these ingredients as well to pull the macros and everything. It’s just not the same.
[Richard Sprague]: Yeah, and I think with labelling, in some ways it may be a dis-service that governments around the world force companies to put the labels on because it gives this false sense of security on your part. That you think, of it’s got sugar in it. Well, what kind of sugar.
[Damien Blenkinsopp]: Exactly.
[Richard Sprague]: And the reality is just way more complicated than they can summarize in a label. And I almost wish that there was instead like a competition among lots of private companies that will compete on the best label that they supply for a particular food.
[Damien Blenkinsopp]: Yeah, because right now everyone hides behind it, basically.
[Richard Sprague]: Exactly, yeah. And in the US it’s particularly bad because we don’t give, it’s not per 100 grams, it’s per serving, whatever that means.
[Damien Blenkinsopp]: I was actually looking at that the other day, and I was like this makes it really hard to do the calculations in my head. Because you always have to have it working on the 100 otherwise you can’t compare.
[Richard Sprague]: And, you know, smarter companies know how to manipulate that. For example, what is it, the little sugar packets that you get for coffee? I guess they’ve arranged it so that they make the size exactly at the cut-off, where now they can say it’s zero calories because it’s like 4.9 calories, but it’s under 5 so they can report it as zero calories.
[Damien Blenkinsopp]: Yeah. There are so many tricks in the food industry. They have mastered the game; they’ve had a while to do it. And I think regulators are never going to be able to solve that, it really has to be transparency coming through because companies want to do it to please their customers. Because regulators, it’s just not their job. You can’t fit a structure that forces people to do it.
[Richard Sprague]: Yeah, and that’s where for the microbiome stuff, as we get more and more companies involved in it and more and more labs that are doing this sort of thing, I do hope that there emerges some sort of independent verification lab, or something.
And I think, was it LabDoor I think that you had on your podcast a while ago is an example of that company that, I love that. They go out and they specifically go and evaluate these things. And it’s independent, and they’re just looking to see kind of, on behalf of consumers, whether you can trust what you’ve got or not.
It’d be neat if there was a similar kind of thing with the microbiome world, wouldn’t it?
[Damien Blenkinsopp]: This is happening, you know the software world has made itself, has been very good at this.
When I think of telecoms, and software, and these IT industries compared to the health industry where it is, there’s a lot of silos in health. And everyone’s got their own lab, and you don’t know if they interrelate, and they don’t, I mean even in the big labs that hospitals have been using for a long time and so long.
And what we really need is a similar structure to what they’ve done in telecoms and software, where you have these big open standards of organization, and everyone gets together and says, we know it’s going to be more valuable for the industry; we know we’re going to make faster progress, and the economics are going to be better, so we’re going to make this.
And maybe it just needs a few people to stand up. So if you’re listening out there, and you’ve got a role in this, go for it please. Because you could add so much value to this industry. People need to start putting things together.
And then I think the other analogy is a lot of integrator kind of companies in software area and the internet now, where things like Zapier and IFFT and all these other apps are relying on all of the rest of them in the ecosystem. And maybe like a conversion app or other ones would add so much value to all of the other things out there.
So I think there’s ways to better integrate these things over time, and it’s going to happen. And there’s plenty of business ideas out there, potentially as well.
[Richard Sprague]: That’s right, yeah. You know, it’d be interesting to look maybe at the history of how, even say like blood testing for standardized.
Because I’m sure they had the same kind of problems in the beginning. Like, how do you decide how to measure Vitamin D, or how do you decide to measure all of this stuff? And it looks like they’ve kind of figured that out. I wonder if some of those same lessons could be applied…
[Damien Blenkinsopp]: Largely, however, I’ve had problems with blood tests in terms of variants. In particular between different countries.
So I was in Spain at one time trying to get labs, and I actually left the country because I gave up completely. Because the data wasn’t correlating with stuff I’d seen in the US and the UK and stuff. So I think there’s still, there is interlab…
[Richard Sprague]: Maybe it’s more complicated than I think.
[Damien Blenkinsopp]: I think there’s still…
[Richard Sprague]: Yeah, a lot of these things, the more you dig into the details the more you realize how messy it all is.
[Damien Blenkinsopp]: Yeah. It’s a crazy world we live in. And this is something you’re constantly working on. You still work on this stuff, do you? Or have you kind of moved on a bit?
[Richard Sprague]: I do. I mean, [most of my situation] right now is I’ve got so much data that I’m spending a little bit more time trying to do the analysis of the data. It is kind of cool though, because every time, lots of people send me their samples and ask me what I think, and every time that somebody sends me a new sample, I get more information.
[Damien Blenkinsopp]: Yeah. Are you offering…
[Richard Sprague]: Yeah. I mean, anybody who wants to, you can send me your uBiome data. I’m happy to look at it and tell you what I think.
I’ll find out little things like, the other day there was a New York Times article about heart disease or something I read this every single day and they’ll mention the particular microbe that was involved. And so I’ll just go look it up and I’ll see, oh, huh. And I log into my computer and I see, okay how does that microbe look in me and what was I doing at the time.
And I’ll find all kinds of interesting correlations. I’ve found things like during travel there are particular microbes that bloom in me. And just we need to understand why, and what is that thing doing, and is it a good thing, a bad thing? I don’t know.
[Damien Blenkinsopp]: I was just also thinking that you’ve travelled a lot, right? You lived in China.
[Richard Sprague]: Yeah, I spent two years in Asia.
[Damien Blenkinsopp]: Right, so we both did that. We both traveled a lot. And I think that influenced our biome a little bit. You found some stuff in there.
[Richard Sprague]: Yeah. One of the things, I mean one of the things I think is really cool is there’s a particular microbe that was identified a few years ago as letting Japanese people, so their digestive systems can handle seaweed, and metabolize seaweed better.
[Damien Blenkinsopp]: And you have it.
[Richard Sprague]: And the study that did this was comparing a lab in Japan versus a lab, I think it was in Saint Louis. And they concluded that North Americans don’t have this, and Japanese people do. And I thought that was pretty cool, but when I looked at my own results I found out I have it too. And that was kind of cool.
In fact, that’s the reason that got me excited about the microbiome, is that it does appear that there are ways that you can change your microbiome.
[Damien Blenkinsopp]: Like living in another country for a while.
[Richard Sprague]: That’s a big one, yeah.
In fact, actually speaking of probiotics. So a lot of people have sent me A/B testing of their probiotics, and one guy sent me, he had three samples. One when he’d been living in the UK, another when he had moved to California and started taking probiotic, and then a month or so later he did another sample. And guess what, you can’t really tell the difference between the two samples taken on the probiotic, but you can tell the difference the sample when he was living in the UK.
(1:52:18) [Damien Blenkinsopp]: Yeah, so if you really want to change your biome, move. So I wonder, I have lived in countless countries. I think my microbiome might be confused, potentially
[Richard Sprague]: Yeah, maybe. But you know what, the microbiome is pretty resilient too. I like looking at, so I compare my father who lives in the Midwest in the United States. And he has kind of stayed in the same place, and that’s where I grew up.
But it’s interesting to look at our microbiomes. I’m essentially a superset of his. So, whatever microbiome I inherited as a result of living in his household for the first however many years of my life, and eating kind of, we have similar tastes in food, and similar diets to this day. But yet I have a superset of his microbiome, because I’ve lived all over the place and he hasn’t.
[Damien Blenkinsopp]: Yeah, exactly.
[Richard Sprague]: And again, that’s kind of a neat thing to know that we do have some influence over how this whole thing turns out.
I run into a lot of people who ask me questions about, like what can I do to change something. And that’s a big one, geography. But there are a lot of things that people don’t necessarily think about, either.
And a big one that I always tell people is about fasting. That’s a fast and easy way to change your microbiome that a lot of people just don’t do. It’s surprising how often you’ll run into somebody who, if you ask them when’s the last time you went a full 24 hours without eating food?
[Damien Blenkinsopp]: Have you seen samples before and after from fasting? I mean, I’m into a lot of fasting so I’d be interested to [know].
[Richard Sprague]: No, I haven’t. And I would like to be able to see that. I would like to see somebody doing a serious job at fasting.
[Damien Blenkinsopp]: I can tell you you need to nurture it back to life after a 10 day fast with fibers. Actually with fibers and stuff, I tried to eat other things, but I was like, it just doesn’t work. So you have to kind of feed it, like I juiced fibers, basically, like vegetables, and actually added some fibers in order to kind of get myself back to normal.
[Richard Sprague]: Yeah. The reason why it’s been hard for me to test this, I mean I do fast occasionally, but it’s hard to test it because when you don’t eat anything, usually you don’t produce anything either. And so.
[Damien Blenkinsopp]: Well, I can tell you a way after a fast to generate poop, just liquid poop, very fast. If you just take fats, that’s not a good idea after fast of five days or so. So that would generate a result quite quickly, but I don’t know what you’d get. It might be completed biased. Yeah, it would be completely biased.
[Richard Sprague]: It would be biased. Yeah.
[Damien Blenkinsopp]: But the other things is, so the solution I found is actually juicing. So if you juice fibers in plants and stuff, and have that as your first couple of drinks, you should after the fast be able to poop quite quickly.
[Richard Sprague]: Yeah, it’s just, you’ll poop something differently than, we don’t know what’s going on in your microbiome before that happens.
I mean, I have tested my skin microbiome extensively like before and after going camping, let’s say. Where I’ll go for several days without a shower to see what happens. And there’s a difference; it’s noticeable. I assume the same thing is happening in the gut microbiome.
[Damien Blenkinsopp]: Yeah. Okay.
[Richard Sprague]: But when I run into people who have some kind of gut issue, that’s one of the first things I suggest is just give it a shot. Because I have talked to people who will say that, you yourself can comment on how fasting does make a difference.
[Damien Blenkinsopp]: Yeah, and that goes back, I always like to quote Valter Longo’s work, where he’s actually got a book out now. But I had my episode on the fast-mimicking diet. Anyone who’s got some weird, chronic issue and that no one knows how to solve it, the cycling of fasting just could be an interesting tool.
[Richard Sprague]: That’s right, and it’s worth tying.
(1:55:42) [Damien Blenkinsopp]: Yeah. Okay, so let’s learn a bit more. Where could someone learn more if they wanted to go an investigate this stuff? Where would you tell people to go and learn more about the microbiome? If they found this whole discussion really fascinating and they want to learn more about the labs and everything.
[Richard Sprague]: Yeah. Where I would start with is, and you can put up a link to it, is I’ve written a post on Medium where I’ve listed my favorite ten books about the microbiome. And that’s what I would look at.
But the number one book I think is Rob Knight’s book about, it’s written a couple of years ago but it’s a great summary; it’s relatively easy and quick to read. It will tell you a lot of the different things that you need to look at. But I do try to read just about every mainstream book that comes out about the microbiome.
And I’ve selected the 10 that I think
[Damien Blenkinsopp]: There’s quite a few coming out now.
[Richard Sprague]: There are a lot of them, yeah. And a lot of them are really excellent. So take a look at my top 10 list. And I’ve tried to keep that up-to-date of the ones that I think are particularly good.
(1:56:32) [Damien Blenkinsopp]: Excellent, excellent. What are the best ways for people to connect with you and learn more about what you’re up to and your work?
[Richard Sprague]: Well the best way is to look at my Twitter handle, just @Sprague. I try to post something pretty regularly. And people are welcome to contact me there. You can also look at my website, richardsprague.com, just my personal website where I kind of post things as they come along.
[Damien Blenkinsopp]: Right. You’ve got your blog over there, right.
[Richard Sprague]: Yeah.
(1:56:57) [Damien Blenkinsopp]: Right, now who besides yourself would you recommend to learn more about the microbiome? Who would be your go to, like your favorite people…
[Richard Sprague]: The favorite person I have is Elizabeth Bik, who on Twitter is @microbiomedigest. And she’s one of the smartest microbiome scientists I know, and she’s very prolific on twitter. She reads all these publications, and she will let you know the ones that matter. So that’s the one I would recommend for that.
[Damien Blenkinsopp]: Wow. Excellent. Is there anyone else?
[Richard Sprague]: A lot of them are the ones that you’ve already featured on your program. Obviously Rob Knight, Eran Segal from Day Two. Those are all good people, that I trust their science and always eager to hear what next thing they’re going to come out with.
(1:57:37) [Damien Blenkinsopp]: Excellent, awesome. Thank you for that. Okay, let’s talk a bit about you. What is your personal approach to improving your body and user tracking? And this is not just microbiome but really anything? Including microbiome.
[Richard Sprague]: Yeah, I’ve been a quantified self-tracker for a long time.
I track my daily amounts of sleep. I track a lot of the main foods that I eat. I don’t do it as rigorously as I’ve done in the past; so like a lot of us there have been times in the past where I rigorously checked. I used to have a Zeo device that I slept with, and I could tell you for years exactly how much REM sleep I had.
And I tracked my activity. Not so much now, I don’t carry a Fitbit or anything, but from time to time I’ll look at just… Because I’ve got such food baselines in the past. If I’m going to make a major change I’ll track myself again.
But the number one thing, I mean I hate to keep on harping on this, but I track my microbiome. I think that’s really fascinating. And it’s something I recommend people, even if you’re not going to track it every day track it once. Get a baseline, and see how it is, and I think you’ll learn a lot.
[Damien Blenkinsopp]: And so what are the things you’ve stuck with now? What are you going to do the next month, or the next three months?
[Richard Sprague]: Well I am interested now in, I’ve been interested in fermented food. One of the things that I discovered from tracking my amount [unclear], power of kefir, because it’s one of the few things that I’ve noticed makes a real, noticeable difference in the microbiome.
And I’m doing a couple of experiments on myself just to see… I’ve noticed a couple of microbes that I did not have when I was before I started drinking kefir and that I have now. One of which is associated with recovery from Crohn’s Disease. So it seems like it’s probably an important microbe.
And I’d like to find out more ones like that. So I’m constantly on the lookout for new kinds of…
[Damien Blenkinsopp]: That’s interesting, and I may be able to help you with that one because I went for a kefir about a year of kefir daily, and I was doing the uBiome test during that period. So there might be in there.
[Richard Sprague]: Oh, interesting. So the data that’s, your uBiome data would include the kefir drink?
[Damien Blenkinsopp]: It would be around it. I think it would be either side of it.
[Richard Sprague]: No, I’ll take a look, because it would be interesting to look to see if you’ve got the microbes that I found in mine.
[Damien Blenkinsopp]: Yeah, that’s what would be interesting, because the first test probably wouldn’t have anything, and then maybe the last test would.
[Richard Sprague]: Yeah. I’m especially interested in traditional, both traditional foods and traditional medicines, because I think that’s an under explored area for finding new interesting microbial solutions to things.
Chinese medicine and Indian Ayurvedic medicine, they have a lot of things that to Western eyes look kind of weird. But if you look at it from the point of the microbiome, suddenly you have a vocabulary now to talk about something in more scientific terms. And I’m really interested in that.
Somebody told me about this, there’s some droplets that apparently Indian mothers give their babies when the babies have colic. And I bet that’s a microbial thing; it probably affects the microbiome.
You know, there’s just little things like that that happen all the time.
[Damien Blenkinsopp]: Right. And before we used to say, there’s no way they can do anything. But as we add these new layers of science on, we start saying actually there’s a potential mechanism there.
[Richard Sprague]: Yeah. And when people have tested some of this stuff, ìscientificallyî, when you look at the details of how they test it, a lot of the times it’ll be something where they, there was some kind of Chinese medicine and somebody will say well let’s bring some people into the lab here in California and let’s give some of them this and some of them that.
Well it’s different conditions than it is when it was administered by a barefoot doctor in rural China, where there are microbes all over the place that are affecting the results. You’re not necessarily comparing apples to apples.
So I think there’s probably a lot of things like that in traditional medicine or food that have a bigger, positive effect than we know. And it’s the kind of thing I wish I knew more about.
[Damien Blenkinsopp]: Cool, very interesting. What I realized now actually is, what kind of insights have you got about your biology from your quantification? And have they led to any changes in behaviors or any actions that you’ve taken? So actually, you know, changes in your life you’ve made.
[Richard Sprague]: Yeah. I would say that I’m pretty healthy. So I’ve not had any real issues that I’ve been concerned about. And so that makes me a little bit, I’m kind of odd. A lot of people who are involved in the microbiome, they have some kind of story about their journey trying to recover from something.
So I don’t really have that. But that also makes me, I think, an interesting case because I’m able to look and see over time how my health as shifted as I get older, and how different things. One of the things that I’m intrigued right now about in particular is sleep.
I’ve always been a reasonably good sleeper, but I get less sleep than a lot of people; I average at around 6.5 hours, and I have for decades. I’m interested in getting better and deeper sleep.
I have found a relationship with potato starch, is one thing that we’ve talked about before, that some people use that as a way to increase the amount of bifidobacterium in their body. It’s something that I would try, if you know somebody who’s having trouble sleeping, that’s one thing to look at.
[Damien Blenkinsopp]: Yeah. Okay, so this is a bit random, but I’ve been working on my sleep for quite a while. I’ve really got to do a full episode on this kind of stuff. And I’ve, like you, had actually worse. At times I get 4 hours sleep, 4.5 hours sleep, and it was very difficult to stay asleep; I can get to sleep, but I can’t basically stay asleep.
So there’s two things that I’ve done that, among all the others, which I think… actually three things. The first is get one of these. So I’m showing Richard a SAD light, 10,000 LUX. SAD Light.
And you put it on and I got this from a Parkinson’s study, because they have problems with sleep as well. And when they showed this, they basically put this on for two hours in the morning.
So it’s basically simulating strong sunlight, right? And you put it next to your desk or something, and you get that. And I’ve found that helps. I think, potentially what’s going on in the mechanism is it’s resetting your sleep cycle. Because we’re not getting enough light; we’re indoors all day, we’re not getting enough light and stuff.
So that seemed to make quite a bit of difference. And the other thing is, which
[Richard Sprague]: So you just turn this light on in the morning?
[Damien Blenkinsopp]: As soon as I get up, I walk into the… I find it’s actually to wake me up as well, better than coffee. Sometimes I’ve forgotten to have my coffee because it’s already done the job, basically.
[Richard Sprague]: Yeah, interesting.
[Damien Blenkinsopp]: So I really
[Richard Sprague]: You just turn it on in the morning, and the rest of the day you turn it off and just live your day?
[Damien Blenkinsopp]:Yeah. And I love this thing.
And I’ve tracked the data and stuff, but I still, I’m still tracking. I got the Oura, which isn’t the best. But I think duration’s not so bad. So I’ve been tracking that over [a longtime]. I’m still kind of waiting to see the results on it.
The other thing is, and this is most people aren’t going to like this, is going to bed really early. And so I started to go to bed, I now, like first of all I said I’m going to get to bed by 11pm. Right?
Because I noticed it seemed that in my Oura data and everything I was like sleeping a longer duration if I got to bed earlier. So that worked a bit. I pulled it back to 10. Worked a bit better. Pulled it back to 9, I’m having 7 hours, 7.5 hours consistently every night, which I’ve never done in my whole life.
And I don’t know why it is. But I can give you like reference of celebrities and people who do this. There’s a lot of people out there that go to bed at 9 and get up at 4. I get a lot more work done as well, now. And I feel much better, but it is a bit of a lifestyle. Most people don’t want to fit in with it.
[Richard Sprague]: Yeah. And that’s interesting that you say that. So are you taking supplements or doing anything special to improve sleep, or just
[Damien Blenkinsopp]: I’ve taken lots of supplements. The only one I still take is glycerin. There’s some studies showing that that helps to reduce night wakings, which is…. So that I do stick with.
In another one of my companies we actually recommend it to anyone, and have them doing it when they have sleep issues and sleep interruptions and that. And it seems to be working consistently across those people.
[Richard Sprague]: Interesting. Yeah, interesting. Yeah, I really miss my Zeo, because before they went out of business that was far and beyond the best way to track your sleep.
[Damien Blenkinsopp]:Everyone misses them.
Well I’m hoping the Next Door is going to be more accurate as well. So that’s coming out, due for delivery in April I think.
[Richard Sprague]: Yeah, it’s hard to see how anything’s going to beat looking at the brain waves, which is what Zeo did.
(2:05:41) [Damien Blenkinsopp]: Okay, right. This is quite an important thing. If you were to recommend one experiment someone should try to improve their body, health, performance, longevity, anything like that, with the biggest payoff, what would that be and how should they track it so they can understand that payoff, and that it’s actually happening for them?
[Richard Sprague]: Yeah. Again, I would look at the microbiome. And probably the number one thing that I see that people could improve with their microbiome is their bifidobacterium levels.
And that’s the thing that, I know you know, it’s associated with sleep, and with serotonin levels. And so just an overall mental stability, all those sorts of things. And what I found in my, looking at lots and lots of samples is that people who don’t have any bifidobacterium, they almost always have some kind of problem.
So the number one thing that I would say for people who are interested in this is to test yourself, see what your bifidobacterium levels are, and then look at different ways to be able to increase it and improve it.
[Damien Blenkinsopp]: Have you got any ideas on what might work?
[Richard Sprague]: Unfortunately, I don’t have really good ideas that work for everyone, but I would start with things like, you can try potato starch which is, if you eat it raw it is known that it’s a particular type of resistant starch that feeds bifidobacterium and it’ll make it through your digestive system. You can try that.
For some people beans work, as I’ve said with my example of going to New Orleans. And then I would test myself in a couple weeks and see if I got any bifidobacterium in me. And I think that’s like the number one thing that I would recommend for people to look at is the bifidobacterium levels, and see what works for changing that in you.
[Damien Blenkinsopp]: Yeah, so that’s a good one. I had non-existent bifidobacterium when I started doing uBiome, but now it pops up in all my tests. So, unfortunately, I can’t say what I did, because I did many different things over that period. But it’s definitely possible.
[Richard Sprague]: Yeah, that’s good news.
[Damien Blenkinsopp]: So that’s good news.
Well, Richard this has been a great discussion. We’ve gone all over the topic, and it’s really great to catch up with you and talk about all this stuff. So thank you for your time.
[Richard Sprague]: No, thank you. It’s always a pleasure talking to you. Damien, you have so many things that you know about, and we’re kind of kindred spirits on this whole quantified journey. So, thanks a lot, it was great talking to you.